Figures & data
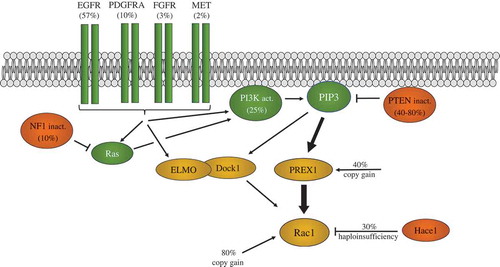
Figure 1. Rac mRNA expression in glioblastoma. (a). mRNA expression levels for Rac1, Rac2, Rac3 and RhoG in the TCGA 2013 glioblastoma database were analyzed using cBioPortal. RSEM normalized data are shown. (b). Rac1 mRNA levels in cases with copy gain (n = 110, 80% of cases) compared to cases without copy gain (n = 27). (c). Frequency of Rac1b splicing in TCGA database samples was analyzed using TCGA SpliceSeq in breast invasive carcinoma, colon adenocarcinoma, lung adenocarcinoma and glioblastoma (GBM). Graphs were generated using SigmaPlot software. Box plots show the median, 10th, 25th, 75th and 90th percentiles. * indicates p < 0.05 by the Mann-Whitney Rank Sum test
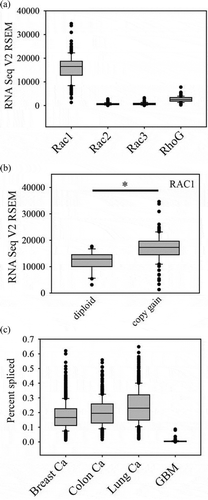
Figure 2. Rac GEF and HACE1 mRNA expression in glioblastoma. (a). Expression of Rac GEFs in glioblastoma. (b). PREX1 mRNA levels in cases with copy gain (n = 52, 37% of cases) compared to cases without copy gain (n = 87). C. HACE1 mRNA levels are decreased in cases with copy loss (n = 41, 30% of cases) compared to cases without copy loss (n = 96). * indicates p < 0.05 by the Mann-Whitney Rank Sum test
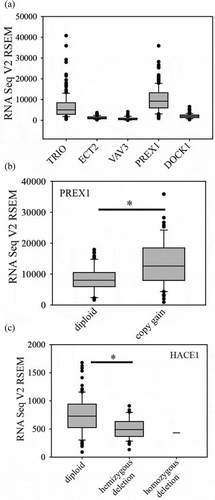
Figure 3. Glioblastoma genetic alterations promoting Rac1 activation. The Epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (a) (PDGFRA), the fibroblast growth factor receptor (FGFR) and the MET receptor (MET) genes are frequently amplified and/or mutated in glioblastoma. This can lead to activation of Rac1 via either PREX1 or ELMO/Dock1 Rac GEFs. PREX1 is directly activated by PIP3 binding, while ELMO/Dock1 requires PIP3 binding as part of its activation mechanism. High PREX1 mRNA and protein levels in glioblastoma suggest it may be the predominant Rac GEF used. PTEN inactivation by copy loss and/or mutation, as well as PI 3-kinase mutational activation, also promotes Rac1 activation via these Rac GEFs. NF1 mutation leads to Ras activation, which can also activate PI 3-kinase and Rac1. Copy gains of RAC1, and PREX1, as well as HACE1 copy loss, may also influence the extent of Rac1 activation. Frequencies of alterations in glioblastoma cases are shown in brackets