Figures & data
Table 1. Clinical characteristics of 1506 patients tested for KRAS status
Table 2. KRAS mutations spectrum in 670 colorectal cancers
Table 3. Correlation of KRAS mutation status with clinical features
Figure 1. Electropherogram for KRAS mutants. Tissue DNA from the patient with colorectal cancer were amplified and cloned for sequencing analysis. Two novel in-flame insertions (10G11 and 11GA12) in exon 2 of KRAS gene were identified.
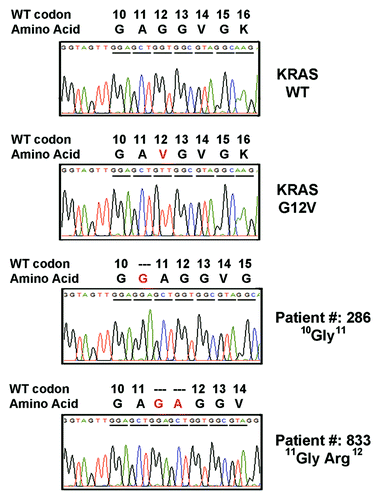
Figure 2.KRAS insertion mutants activated RAS signaling by enhancing cellular accumulation of active RAS (RAS-GTP) and activating p-ERK. NIH3T3 and 293FT cells were transfected with KRAS mutants, and RAS-GTP protein in the cell extract were immunoprecipitated with agarose beads containing Ras binding domain of Raf-1. Protein levels in both whole cell extracts (pan-RAS and pERK) and precipitated samples (RAS-GTP) were analyzed by western blot analysis as indicated. Representative results from 3 independent experiments were shown.
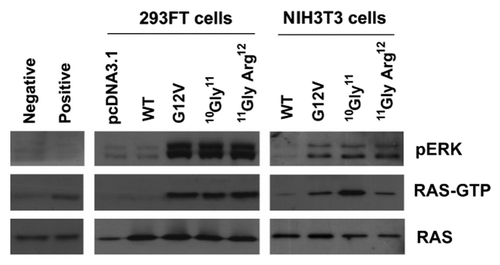
Figure 3.KRAS insertion mutants promoted anchorage-independent growth in soft agar. NIH3T3 cells stably transfected with pcDNA3.1 empty vector (EV), wild-type KRAS (WT), G12V KRAS mutant (G12V), 10G11 and 11GA12 mutants were cultured in soft agar for analysis. Representative microscopic pictures of colony from each transfectant were taken (Magnification, 400×). The number of colony in each transfectant was plot in the bar chart and the results shown were mean and standard deviation from three independent experiments. The P value of < 0.05 and < 0.001 were denoted as * and ** respectively.
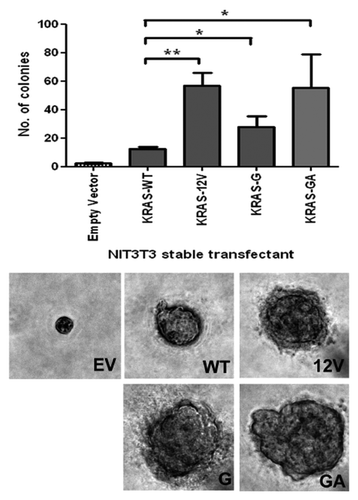
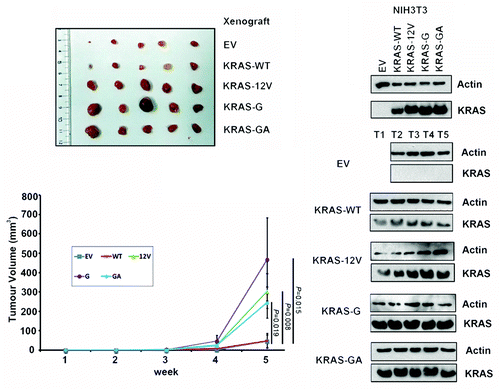
Figure 4.KRAS insertion mutants promoted in vivo growth of NIH3T3 cells. In vivo tumorgenic assay in nude mice showed that tumors formed in the sites implanted with NIH3T3 cells expressing KRAS mutants (G12V, 10G11, or 11GA12) were consistently larger than that implanted with wild-type KRAS (WT) and empty vector (EV) controls. By western blotting, the expression of KRAS protein in the NIH3T3 transfectants and tumors dissected from the xenografts (T1–T5) was detected.