Abstract
Despite advances in modern technologies, various foodborne outbreaks have continuously threatened the food safety. The overuse of and abuse/misuse of antibiotics have escalated this threat due to the prevalence of multidrug-resistant (MDR) pathogens. Therefore, the development of new methodologies for controlling microbial contamination is extremely important to ensure the food safety. As an alternative to antibiotics, bacteriophages(phages) and derived endolysins have been proposed as novel, effective, and safe antimicrobial agents and applied for the prevention and/or eradication of bacterial contaminants even in foods and food processing facilities. In this review, we describe recent genetic and protein engineering tools for phages and endolysins. The major aim of engineering is to overcome limitations such as a narrow host range, low antimicrobial activity, and low stability of phages and endolysins. Phage engineering also aims to deter the emergence of phage resistance. In the case of endolysin engineering, enhanced antibacterial ability against Gram-negative and Gram-positive bacteria is another important goal. Here, we summarize the successful studies of phages and endolysins treatment in different types of food. Moreover, this review highlights the recent advances in engineering techniques for phages and endolysins, discusses existing challenges, and suggests technical opportunities for further development, especially in terms of antimicrobial agents in the food industry.
Introduction
Safe food is a fundamental requirement in everyday life. However, food can readily be contaminated by pathogenic microorganisms, posing a critical health threat to the public and a substantial economic burden on society. Furthermore, the recent globalization of the food market and lengthy food supply chain for ready-to-eat (RTE) products may increase the likelihood of cross-contamination with/or by microbial pathogens (Lineback et al. Citation2009). Despite efforts to control microbial contamination in various foods, including hazard analysis and critical control points (HACCP), and good manufacturing practices (GMP), the reported number of foodborne illnesses and their economic burdens have increased globally over the past decade (WHO Citation2020). The World Health Organization (WHO) stated that more than 250 known and unknown diseases are transmitted to humans by consumption of contaminated foods, causing serious foodborne illness (requiring hospitalization) of approximately 600 million people and more than 420,000 deaths annually (WHO Citation2020). The financial impact of this problem, including direct healthcare costs and indirect loss of productivity costs, is estimated to exceed $110 billion on a global scale (WHO Citation2020). Therefore, maintaining food quality and safety at an elevated level is an important challenge in the food industry.
As penicillin has demonstrated its potent efficacy in the human body since the 1940s, many countries worldwide have used antibiotics as therapeutic agents in humans and farm animals (CDC Citation2020). However, the overuse and misuse of antibiotics in human clinics and agriculture has caused the emergence and spread of multidrug-resistant (MDR) bacteria (English and Gaur Citation2010). In agriculture, antibiotics have been treated to farm animals such as pigs, cows, and chickens in order to prevent infections and improve growth and production since the 1940s. In addition, the increased demand for animal protein instigated the overuse and misuse of antibiotics in animal-derived products (Manyi-Loh et al. Citation2018). Antibiotic-resistant bacteria or MDR pathogens are currently one of the greatest threats to humankind and the most widespread global public health concern. The WHO has carried forward the Global Antimicrobial Resistance Surveillance (GLASS) project in 2015 to defeat the threat of antibiotic resistance (WHO Citation2016) and stressed the severity of 12 species of antibiotic-resistant bacteria as ‘global priority pathogens (GPP)’ in all international regions in 2017 (WHO 2017). According to the Centers for Disease Control and Prevention (CDC)’s report of antibiotic resistance threats in the United States (CDC Citation2019), approximately 2.8 million people are infected by antibiotic-resistant bacteria in the U.S. each year, and more than 35,000 people die annually from infections as a result. Additionally, the estimated national cost to treat infections caused by six MDR germs is substantial – more than $4.6 billion annually (CDC Citation2019). Agriculture accounts for approximately 80% of the total annual antibiotic use in the USA (OECD. Citation2018). Thus, the extensive use of antibiotics makes animals, food, and agriculture critical sectors in the fight against antimicrobial resistance (AMR). Hence, novel antimicrobials for the biocontrol of foodborne pathogens with different modes of action of antibiotics are urgently needed.
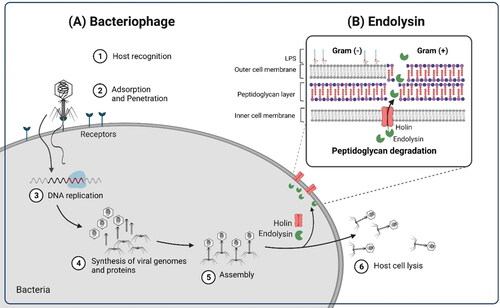
Bacteriophages (phages) and their derived enzymes, endolysins, and depolymerases have the potential to be promising intervention tools for this purpose () (Lin, Koskella, and Lin Citation2017, Romero-Calle et al. Citation2019). Phages are the most abundant and widespread biological entities in the biosphere, including foods and environments, with an estimated population of 1031 (Brüssow and Hendrix Citation2002; Dion, Oechslin, and Moineau Citation2020). Phages are viruses that can infect a highly specific set of bacterial species or strains and do not affect and alter commensal microflora (Cieplak et al. Citation2018). Pathogen-specific antimicrobials are incredibly beneficial, considering the undesirable side effects because of antibiotic administration such as reduced diversity of the beneficial microbiota (e.g., gut dysbiosis) (Francino Citation2016). Moreover, the feasibility of simple, fast, and relatively lower cost phage production using the self-replicating nature of phages is an additional advantage for their use as antimicrobials (Azeredo and Sutherland Citation2008). Endolysins, a specific type of peptidoglycan hydrolase (PGH), enzymatically degrade the peptidoglycan layer of specific bacteria and have several properties as a good antimicrobial (Borysowski, Weber-Dabrowska, and Górski Citation2006; Oliveira et al. Citation2013). These enzymes are produced from phages at the final stages of the multiplication cycle and cleave the specific site of the peptidoglycan layer to degrade the host cell, resulting in the release of progeny virions (Channabasappa et al. Citation2018; Loessner Citation2005). In particular, endolysins can rapidly kill host bacteria by degrading their cell walls within minutes or even seconds (Loessner Citation2005), illustrated by the fact that some endolysins eliminated 107 Streptococcus pyogenes bacteria to undetectable levels 10 s after treatment (Fischetti Citation2003). Moreover, endolysin-resistant bacteria have rarely been reported to date (Fischetti Citation2005).
Figure 1. Mechanisms of action of (A) bacteriophage and (B) endolysin. The figure was created with Biorender (http://biorender.com).
To be used as food antimicrobial agents, candidates should satisfy several properties: (i) adequate host range, (ii) harmless to human consumption, (iii) characteristics that do not affect taste and organoleptic properties of food, and (iv) maintenance of antimicrobial efficacy under extreme food processing conditions (Kumar, Ravishankar, and Juneja Citation2017). Although natural phages and endolysins are eco-friendly antimicrobials that meet some of the requirements mentioned above, they have several limitations (e.g., narrow host spectrum, phage-resistance problem, and low stability) that should be addressed before they can be used as ideal antimicrobial agents for realistic application to foodborne pathogens. Recently, the increased registration of phage genome resources in public databases and advances in synthetic molecular biology toolkits have offered attractive options for engineering native phages and endolysins. These genetic and protein engineering approaches may be possible solutions to overcome the disadvantageous aspects of native phages and endolysins, opening up the next chapter for applying phage-derived antibacterial agents with maximized advantages in the food industry.
In this review, we aim to predict the potential of engineered phages and endolysins, especially in terms of novel antimicrobial agents against foodborne pathogens on food and food contact surfaces. We introduce an up-to-date overview of the engineering strategies for phages and endolysins and discuss the recent advances in engineering techniques for their application in the food industry. Furthermore, we present extant obstacles that still need to be addressed and propose technical approaches to overcome these drawbacks in the future.
Bacteriophages (phages): Abundant and potential weapons for food safety
Compared to conventional antibiotics, phages have unique characteristics for being applied as alternative antimicrobial agents: no toxic effect on human or animal cells, lower costs for development and production than novel antibiotics, and self-dosing (Matsuzaki et al. Citation2005; Meaden and Koskella Citation2013). Phages are broadly divided into two groups depending on their life cycle: virulent phage (lytic cycle) and temperate phage (lysogenic cycle) (Kutter and Sulakvelidze Citation2004, Kasman and Porter Citation2020). Virulent phages typically lead to cell death immediately after mature phage particles are produced, minimizing the possibility of transducing virulence or antibiotic resistance genes (Hassan et al. Citation2021). Thus, virulent phages are suitable antimicrobial agents to control foodborne pathogens and spoilage organisms in pre- and post-harvest foods.
The concept of ‘phage therapy’ was first suggested by Felix d’Herelle in 1917 for therapeutic and prophylactic applications. Due to the great potential of phages against infectious diseases, many countries have conducted diverse types of practical research, especially in Eastern Europe and the Soviet Union (Fruciano and Bourne Citation2007). However, research on phage therapy in Western countries was discontinued after the introduction of antibiotics such as penicillin in the 1940s. The dark age was because chemical antibiotics cured illnesses more efficiently than other antimicrobial agents (Dublanchet and Bourne Citation2007). After the emergence of MDR bacteria in the 1980s, phage therapy was reconsidered as an interesting method to combat MDR bacteria. In 2006, the phage-based product ListShield (Intralytix, Inc., Baltimore, MD, USA) was first approved by the United States Food and Drug Administration (FDA) and the U.S. Department of Agriculture (USDA) as a food additive to control Listeria monocytogenes in ready-to-eat food (Bren Citation2007) and permitted as GRAS status by the FDA in 2014. Since then, other phage products such as EcoShield (Intralytix), SalmoFresh (Intralytix), and Agriphage (OmniLytics Inc., Sandy, UT, USA) also have been approved for commercial use in food safety (Moye, Woolston, and Sulakvelidze Citation2018).
Bacteriophage applications in foods
Microbial contamination can occur during both pre-and post-harvest environments or during the manufacturing process because food contains various nutrients and minerals required for bacterial growth. Therefore, the treatment of specific phages can protect food materials from damage caused by bacteria during food production. Many studies have been conducted on biocontrol assays using phages in various foods to control various foodborne pathogens (Połaska and Sokołowska Citation2019), several countries including Canada, the USA, Switzerland, Israel, Australia, and New Zealand have allowed for use of phage therapy in food (Au et al. Citation2021). In this section, we will briefly introduce the cases of phage applications in several types of food matrices: (i) dairy products, (ii) meat, (iii) vegetables or crops, nd (iv) biofilms ().
Table 1. Phage applications in the foods and biofilm.
Dairy products can be easily contaminated at many points along the production chain, even after pasteurization (Montgomery, Haughey, and Elliott Citation2020). According to the CDC, many outbreaks caused by Listeria in cheese have been reported, including the recent case of contaminated queso fresco cheese (CDC Citation2021). To prevent listeriosis in dairy products, PhageGuard Listex (Intralytix) and ListShield, commercial phage products, were administered to artificially contaminated surface-ripened red-smear soft cheese, resulting in a significant reduction of Listeria (Carlton et al. Citation2005; Guenther and Loessner Citation2011; Perera et al. Citation2015). In addition to Listeria, phage treatment can prevent dairy products from contaminating pathogenic Escherichia coli O157:H7 and Staphylococcus aureus (Gharieb et al. Citation2020; Li et al. Citation2021).
Poultry and meat products are consumed worldwide. The CDC has reported many outbreaks caused by Salmonella spp., Campylobacter spp., and Shigella spp. contaminations, which cause illnesses such as salmonellosis, campylobacteriosis, and shigellosis in poultry and meat (Kim et al. Citation2019; Moye, Woolston, and Sulakvelidze Citation2018; Shin et al. Citation2012). Several phage products have been developed and approved by the FDA to prevent Salmonella contamination in livestock and food. SalmoFresh (Intralytix) treatment by dipping and applying to the surface reduced the populations of Salmonella by up to log 0.9 and 1.2 after a day of storage in chicken breast fillets (Sukumaran et al. Citation2016). Another phage preparation, PhageGuard S (PhageGuard, Wageningen, Netherlands), successfully controlled the Salmonella (1 log reduction) in ground beef (Yeh et al. Citation2018). Besides, the FDA-approved product, ShigaShield (Intralytix), removed artificially contaminated Shigella on deli meat and pre-cooked chicken (Soffer et al. Citation2017).
The consumption of fresh products, including vegetables and fruits, has increased owing to the demand for healthy diets (Żaczek, Weber‐Dąbrowska, and Górski Citation2015). However, contaminations in fresh products occurred for several reasons such as inadequate storage temperature, poor worker hygiene, and use of bio-contaminated equipment (López-Cuevas et al. Citation2019). The FDA announced that foodborne pathogens, such as Salmonella spp., E. coli O157:H7, L. monocytogenes, and Shigella spp. in fresh fruits and vegetables, can cause foodborne illnesses (FDA. Citation2008). Phage products were applied to fresh products as prophylactic agents to prevent microbial contamination, and the US Environmental Protection Agency (EPA) approved the use of phages as biopesticides to control harmful plant pathogens on crops, including tomato, pepper, apple, and citrus (EPA 2007) (Goodridge and Bisha Citation2011). The use of SalmoFresh was efficient for the biocontrol of Salmonella on romaine lettuce and sprouts (Zhang et al. Citation2019). Many studies used Ecoshield (Intralytix) to control the E. coli O157:H7 in tomatoes, broccoli, spinach, and lettuce (Abuladze et al. Citation2008; Sharma et al. Citation2009). Fresh products are stored and displayed at low temperatures to maintain product quality from harvest to consumers. Refrigerated foods are likely to be contaminated with Listeria during processing since Listeria can grow well under cold conditions (Junttila, Niemelä, and Hirn Citation1988). Listshield (Intralytix) and PhageGuard Listex (Intralytix) were applied to the surface of artificially contaminated vegetables or fruits, and effectively reduced the number of Listeria (Leverentz et al. Citation2003, Citation2004).
Biofilms are a major concern in the food industry at any step in the food chain because food spoilage often occurs because of contact with biofilms formed on the surface of contaminated equipment, and hand during processing, packaging, and cooking processes (DeVita et al. Citation2007). Phages can degrade polysaccharides such as a capsule or LPS O-antigen by using hydrolytic enzymes to invade the biofilm structure, which results in the elimination of biofilms (Abedon et al. Citation2021). Listex P100 treatment reduced the L. monocytogenes population attached or grown on stainless steel surfaces by log 3.5–5.4 CFU/cm2 (Soni and Nannapaneni Citation2010). SalmoFresh and SalmoLyse protected stainless and glass surfaces from Salmonella contamination by more than 99% (2.1–4.3 log) (Woolston et al. Citation2013).
Limitations of phages as antimicrobial agents
Phages have several limitations for food antimicrobial agents in the food industry, although many phage applications have been reported. First, phages can infect a limited number of bacterial strains within the same or closely related species (Abdelsattar et al. Citation2021; Vikram, Woolston, and Sulakvelidze Citation2020). This is due to various factors, including specific binding between receptor binding proteins (RBPs) and bacterial host receptors, superinfection exclusion, and diverse phage-resistant mechanisms (Abdelsattar et al. Citation2021; Ross, Ward, and Hyman Citation2016). Although various phages are isolated by labor-intensive and time-consuming experiments, most phages have a narrow host range. Thus, not all isolated phages from nature can be employed as effective antimicrobial agents (Loc-Carrillo and Abedon Citation2011). Second, the emergence of phage-resistant bacteria poses a potential risk. Bacteria can resist phage infection through spontaneous mutations in genes associated with the biosynthesis of phage receptors, which typically leads to decreased fitness and reduced virulence (León and Bastías Citation2015). However, bacteria such as Salmonella could develop transient phage resistance through phase-variable O-antigen glucosylation (Kim and Ryu Citation2012), suggesting that phage resistance is not permanently maintained and does not always lead to loss of function of phage receptors. Therefore, phage-resistant bacteria that remain in food matrices can cause diseases. Third, phage structures, made of head and tail, are composed of proteins and are affected by external factors such as temperature, acidity, and salinity (Jończyk et al. Citation2011). Thus, phage virions can be unstable when applied to food environments because many factors can affect protein stability during food processing (Xu Citation2021). Phages infecting Gram-negative bacteria presented low survivability after 40 min incubation at 62 °C (80/83) (Jurczak-Kurek et al. Citation2016) and twenty-one Salmonella phages were unstable when exposed to high temperatures (70 °C) for 30 min (Park et al. Citation2021). To overcome this limitation, Park et al. (Citation2021) recently suggested a phage isolation method of applying heat treatment (70 °C, 30 min) to environmental samples and showed that 14 thermostable phages efficiently controlled and prevented Salmonella contamination in food models (milk and chicken breast).
Genetic engineering of phages
Some phage products are used as food antimicrobial agents, but their inherent drawbacks need to be overcome to develop more efficient phage-based antimicrobials (Chen et al. Citation2019; Huss and Raman Citation2020). Thus, many studies have recently attempted to improve the properties of phages through synthetic biology and genome engineering. We will introduce how genetic engineering strategies can increase the applicability of native phages ().
Table 2. Published strategies for phage engineering.
Table 2. (Continued)
Homologous recombination
Homologous recombination is one of the most well-studied strategies for genome engineering in bacterial hosts. During the phage infection cycle, gene insertions or deletions can occur via homologous recombination between the linear or circular form of heterologous DNA and the phage genome. It naturally occurs between two DNA molecules with homologous sequences of at least 20 bp in bacteria (Vos Citation2009). The introduction or exchange of the gene encoding tail fiber has been reported to expand the host range of Enterobacteria phage T2 and Pseudomonas phage JG004 (Le et al. Citation2013; Mahichi et al. Citation2009; Yoichi et al. Citation2005), or to change the host spectrum of Acinetobacter baumannii phage ΦAB1 (Lai et al. Citation2016). Host-range determining regions (HRDRs) in T3 phage tail fiber proteins were analyzed and mutated using phagebody libraries, resulting in diverse recombinant phages with altered host ranges and suppression of bacterial growth (Yehl et al. Citation2019). In addition to the host spectrum, the introduction of the depolymerase gene into the phage genome enhanced the antimicrobial activity and biofilm-removing efficacy of phages (Born et al. Citation2017; Lu and Collins Citation2007).
Although various recombinant phages have been constructed, recombination frequencies are very low, from 10−8 to 10−4 (Loessner et al. Citation1996; Mahichi et al. Citation2009), and the screening process was inefficient. Phage lambda proteins (Gam, Exo, and Beta) and RecE/RecT-like proteins have been used in genome engineering of various types of phages targeting E. coli and Salmonella to promote recombination efficiency (Jensen et al. Citation2020; Marinelli et al. Citation2008; Shin et al. Citation2014). One of the adaptive immune systems of bacteria, clustered regularly interspaced short palindromic repeats and associated genes (CRISPR-Cas), was used to counter-select non-edited phage genomes in phage engineering. This strategy has been reported since 2014 and enabled precise gene insertion or substitution at specific loci (Hatoum-Aslan Citation2018). Endoribonucleases, such as Cas3 and Cas9, cleave the foreign nucleic acid binding with the CRISPR RNA (crRNA), and the DNA breaks are repaired by homologous recombination with donor plasmids encoding upstream and downstream regions of the target gene. The desired recombinant phages are selected by the CRISPR-Cas system (Kiro, Shitrit, and Qimron Citation2014). Different classes of the CRISPR-Cas system, including types I-E, II-A, III, and V, were applied to genome engineering of phages targeting foodborne pathogens, such as E. coli phage T7 (Kiro, Shitrit, and Qimron Citation2014), T4 (Dong et al. Citation2021; Duong et al. Citation2020, Tao et al. Citation2017), T5 (Ramirez-Chamorro, Boulanger, and Rossier Citation2021), and Staphylococcus epidermidis phage Andhra and ISP (Bari et al. Citation2017).
Genome assembly/rebooting
Advances in genetic manipulation techniques, such as enzymatic in vitro assembly (Gibson assembly), allowed the rapid synthesis of partial fragments of the phage genome, followed by assembly of the full-length phage genomes (Gibson et al. Citation2009). Kilcher et al. (Citation2018) reported a platform technology that can rapidly construct recombinant phage targeting Gram-positive bacteria by employing Listeria L-form cells without a selection process. Listeria L-form cells were transfected with the assembled whole phage genome by Gibson assembly, successively reactivating the genomes of phages infecting Listeria, Bacillus, and Staphylococcus. This approach was applied to find which T3 phage genes are associated with survivability under high-temperature conditions (Favor et al. Citation2020), mutations within each gene (T3p37 and T3p45) caused enhanced stability.
As a different method for genome rebooting, the yeast-based platform was used for stable recombination of the phage genome (Pires et al. Citation2016). The fragmented full-length genome of phage phiX174 was ligated with linear yeast artificial chromosome (YAC) molecules using transformation-associated recombination (TAR cloning) and electroporated into E. coli cells to recover phage particles (Jaschke et al. Citation2012). Ando et al. (Citation2015) constructed a high-throughput strategy to reboot several T7-like phages that infect E. coli, Klebsiella, and Yersinia bacteria. The authors modulated the phage host range by inserting the tail fiber genes of Klebsiella phage K11 into the genome of the T7 phage. Furthermore, three nucleotide mutations were introduced to the T3 gene 17 so that the T3 phage encodes the same tail fiber as Yersinia phage R. As a result, the synthetic T3 phage could infect both E. coli and Yersinia. Larger phage genomes, such as Salmonella phage FelixO1, were also recovered using this method (Timothy Kuan Ta Lu et al. Citation2011). Recently, Latka et al. (Citation2021) recombined the structural module of the RBPs to switch the capsule serotype specificity and host range of Klebsiella phages. The domains of RBPs with depolymerase activity were randomly assembled using the VersaTile method, and recombinant phages showed altered serotype specificity according to swapped RBPs. Although these approaches can synthesize stable phage genomes in yeast, they have a limitation in that host bacteria should be competent to uptake large DNA molecules.
Cell-free genome synthesis
Many studies have revealed that the whole phage genome can be synthesized using an in vitro assembly protocol and reactivated as recombinant phages in the host bacteria. The assembled phage genome has to be transformed into host bacteria for phage rebooting; however, the transformation efficiency of most bacteria with large DNA molecules, especially Gram-positive bacteria, is very low. To overcome this obstacle, a cell-free transcription and translation (TX-TL) system that mimics the cytoplasmic environment of E. coli was developed by Jewett and Swartz (Citation2004) and was first applied to produce MS2 phage coat protein virus-like particles (MS2 VLPs) (Bundy, Franciszkowicz, and Swartz Citation2008). Furthermore, E. coli phage T7, phiX174, and MS2 were successfully synthesized in the test tube using the optimized TX-TL system with thioredoxin and dNTPs (Garamella et al. Citation2016; Shin et al. Citation2012), and the authors presented a complete synthesis of the T4 phage, which has a large genome (168 kbp) with a high yield (Rustad et al. Citation2018). Furthermore, they have optimized this system’s transcription and translation rate for efficient phage or fluorescence protein synthesis (Garenne et al. Citation2021). Another study used chemically synthesized oligonucleotides for de novo phage genome synthesis, resulting in the production of the Acinetobacter phage AP205 (4268 bp) at a low cost ($0.0137/bp) and low error rate (Yeom et al. Citation2020).
Endolysins: potential antimicrobials to replace antibiotics
In light of the challenges posed by existing phage applications, endolysins are attracting more attention from practitioners and the industry, various endolysin and its application in food have been reported from many countries such as European Union, the USA, and East Asia. They are effective in the lysis of Gram-positive bacteria when treating purified endolysins outside the cells (Pastagia et al. Citation2013). This exogenous antibacterial effect is more potent against Gram-positive bacteria than against Gram-negative bacteria because the peptidoglycan layer of Gram-positive bacteria is exposed to the exterior medium, facilitating the direct access of endolysins to cell wall carbohydrates and peptidoglycan (Briers et al. Citation2014).
Endolysins targeting Gram-positive bacteria and some endolysins targeting Gram-negative species are characterized by a modular architecture, constituting several domains. Gram-positive endolysins are composed of at least two distinct domains connected by a flexible linker of variable length between (i) the enzymatically active domain (EAD) at the N-terminus and (ii) the cell wall-binding domain (CBD) at the C-terminus (Gerstmans, Criel, and Briers Citation2018). The EADs participate in the breakdown of various bonds in the peptidoglycan, thereby embodying the muralytic action of an endolysin (Loessner Citation2005; Nelson et al. Citation2012). There are at least five different groups of EADs classified according to the specific sites of a peptidoglycan layer at which each type attacks (Borysowski, Weber-Dabrowska, and Górski Citation2006; Nelson et al. Citation2012). Meanwhile, CBDs recognize cell wall-associated ligand molecules and increase the affinity of enzymes to their substrates, conferring host specificity (Schmelcher, Donovan, and Loessner Citation2012a). Generally, the EAD could be up to two domains, and an additional spore-binding domain (SBD) can be added at the end of the CBD (Kong et al. Citation2019). In contrast, Gram-negative endolysins generally possess a globular structure with only EADs (Gerstmans, Criel, and Briers Citation2018).
Endolysin applications in foods
Endolysins are potential antimicrobials in the food industry. Unlike the phages, the use of endolysins in food is regarded to be safer because they do not create gene transfer issues or contribute to the emerging problem of resistant bacteria (Loeffler, Nelson, and Fischetti Citation2001; Pastagia et al. Citation2011; Schuch, Nelson, and Fischetti Citation2002). Indeed, two studies were conducted to identify and isolate strains resistant to endolysins, but no strains have been identified (Fischetti Citation2005; Loeffler, Nelson, and Fischetti Citation2001). Fundamental resistance to these enzymes rarely emerges as it requires mutations in specific bonds in the peptidoglycan, which are evolutionarily conserved and essential (Fischetti Citation2005; Nelson et al. Citation2012; Schmelcher, Donovan, and Loessner Citation2012a). Compared to antibiotics, endolysins possess limited host specificity (Pagan Citation1981), which excludes the risk of killing other beneficial microorganisms in food products. Moreover, rapid lysis of Gram-positive bacteria cells within minutes with a small amount of enzyme is another powerful advantage of endolysin (Loessner Citation2005).
Indeed, the antimicrobial activity of endolysins has been studied by targeting several kinds of food products: (i) dairy products, (ii) meat, (iii) vegetables or crops, and (iv) biofilms (). Dairy products are the most widely studied food type for the efficacy of endolysins. In the dairy manufacturing industry, dairy-borne pathogens, including enterotoxin-producing S. aureus, Shiga toxin-producing E. coli (STEC), L. monocytogenes, and Salmonella species should be controlled (Oliver, Jayarao, and Almeida Citation2005). Various endolysins have shown effective lytic activity against S. aureus in milk (Chang, Kim, and Ryu Citation2017a; García et al. Citation2010a; Obeso et al. Citation2008). In addition, a few endolysins have been tested and analyzed as antimicrobial agents against Clostridium sporogenes in milk (Mayer et al. Citation2010) and, Listeria in dairy products such as milk (Schmelcher, Waldherr, and Loessner Citation2012b), soy milk (Zhang et al. Citation2012), and Queso Fresco cheese (Ibarra-Sánchez, Van Tassell, and Miller Citation2018).
Table 3. Endolysin applications in the foods and biofilm.
Moreover, meat is another important contaminant of foodborne pathogens because animal-derived food has a high nutritional content, high water activity, and neutral pH (Bantawa et al. Citation2018). To combat pathogens in meat or livestock, few studies have determined the bactericidal activity of endolysins in MRSA-contaminated ham (Chang, Kim, and Ryu Citation2017a). Endolysin treatment has also been recommended as a strategy to control pathogenic bacteria in vegetables, avoiding the antibiotic resistance problem. For antimicrobial agents of vegetable products, several endolysins have been suggested as potential antimicrobial agents against L. monocytogenes (Schmelcher, Donovan, and Loessner Citation2012a) in iceberg lettuce. Together with the direct treatment of foods with antimicrobials, the sanitation of biofilms on food-contacting materials in food processing chains is also vital to block cross-contamination of bacteria and guarantee food safety. Studies on endolysin treatment have been performed recently to combat biofilms formed by common biofilm-forming pathogens, including Staphylococcus, Streptococcus, and Listeria. In particular, many staphylococcal endolysins have shown powerful biofilm removal ability against S. aureus and S. epidermidis (Cha et al. Citation2019; Gutiérrez et al. Citation2014; Son et al. Citation2010).
Limitations of endolysins as antimicrobial agents
Despite the effectiveness of endolysins, several limitations remain in the practical use of endolysins in the food industry. Even though there are various endolysins in nature, the identification of novel and powerful endolysins requires prerequisite steps, including phage isolation, propagation, endolysin cloning, and purification (Lee, Son, et al. Citation2021). However, some endolysins show poor expression levels and insolubility (Son et al. Citation2021). Moreover, research on endolysins as food antimicrobial agents is still in its infancy because many applications of endolysins have focused on their preventive and therapeutic efficacy against bacterial infections in animal models (García et al. Citation2010b). Endolysins have variable antibacterial efficiency depending on the food components (proteins, carbohydrates, fat, minerals, and vitamins) and their biochemical factors (temperature, pH, and ionic strength) in foods (Shannon, Radford, and Balamurugan Citation2020). All these factors could affect the protein stability and/or activity of endolysins, alleviating their practical use. Thus, relatively limited scientific studies have confirmed their feasibility in food products (Shannon, Radford, and Balamurugan Citation2020).
Synthetic engineering of endolysins
During co-evolution, phages do not unilaterally annihilate bacteria because the host bacteria are required for phage replication. Thus, we assume that endolysins have not evolved to exhibit maximum activity (Ryu Citation2021). There are many opportunities for engineering endolysins, which can deviate from the major limitations of natural endolysins: (1) low bactericidal activity and a narrow host range in the presence of food components, and (2) inability to kill Gram-negative bacteria effectively. The low catalytic efficiency and narrow host spectrum of endolysins could be redirected based on a key characteristic of endolysins, “modularity” General Gram-positive endolysins have a modular architecture comprising diverse EADs and CBD (Gerstmans, Criel, and Briers Citation2018). Their functional domain units can provide easy and efficient engineering toolkits: (i) domain truncation or deletion, (ii) direct mutagenesis, and (iii) domain swapping method ().
Table 4. Published strategies for endolysin engineering.
Table 4. (Continued)
Truncation or deletion of domains
Truncation or deletion of a domain can modify the antibacterial activity, CBD dependence, and host specificity of endolysins. Cheng and Fischetti (Citation2007) first conducted random mutagenesis of Streptococcus endolysin PlyGBS using mutant strains. Mutated endolysin, which yielded approximately 20-fold improved lytic activity compared to the full-length PlyGBS, was revealed as a truncated endolysin by chance. As a more rational approach, Horgan et al. (Citation2009) also constructed different derivatives of the LysK endolysin of Staphylococcus and revealed that CHAPk (a truncated LysK endolysin) showed better lytic activity against staphylococcal strains than its natural form in vitro. In contrast to previous findings, Low et al. (Citation2011) discovered the increased enzymatic activity of some endolysins upon removing their CBD. They explained this phenomenon in a context in which the CBD dependence of an EAD domain is closely related to its charge. As lipoteichoic acids in the cell wall are negatively charged, a change in the net charge of the catalytic domain from a negative to a positive one may result in increased interaction between the catalytic domain and lipoteichoic acids.
Direct mutagenesis
Direct mutagenesis was conducted to upgrade the endolysins with enhanced lytic activity and even stronger thermostability. Mayer et al. (Citation2011) discovered that direct mutagenesis in conserved residues responsible for a hydrogen bond networking at the backside of the catalytic sites of the amidase domain resulted in the increased lytic activity of mutated CD27L endolysin against several L. monocytogenes serovars. Díez-Martínez et al. (Citation2013) constructed a synthetic endolysin, Cpl-7S, by substituting 15 amino acids into the CBD. This substitution increased the net charge of CBD, thereby inducing better lytic activity. Love et al. (Citation2021) mutated a single residue of a non-conserved hydrophobic core based on the understanding of mutable positions within the LysF1 fold, increasing the thermal stability. Site-directed mutagenesis based on protein structure can be a more rational approach for improving the properties of endolysins.
Domain swapping method
As each domain of endolysins can independently function, domains can be interchanged or recombined with domains of different endolysins to generate chimeric enzymes with desirable properties. Indeed, several studies have demonstrated successful outcomes of these strategies in increasing catalytic efficiency (Díez-Martínez et al. Citation2015; Mao et al. Citation2013; Yang et al. Citation2014) and extending host specificity by either CBD swapping or addition (Becker et al. Citation2009; Yoong et al. Citation2004; Zhou et al. Citation2017). Fusion endolysins with increased lytic activity and thermostability have been successfully applied to food matrices such as milk and poultry. Rodríguez-Rubio et al. (Citation2012) demonstrated that the fusion protein, CHAPSH3b (consisting of the CHAP domain of HydH5 and the SH3b domain of lysostaphin), killed S. aureus to undetectable levels in pasteurized milk after 15 min of treatment at a relatively low concentration (1 μM). Mao et al. (Citation2013) also enhanced the lytic activity of Ply187 in milk by generating a fusion protein of the EAD of Ply187 (Ply187AN) and the CBD of LysK (KSH3b). Swift et al. (Citation2015) generated an engineered endolysin (PlyGVE2CpCWB) by fuzing the amidase domain from the endolysin derived from the thermophilic phage ΦGVE2 to the CBD from the endolysin of Clostridial phage ΦCP26F.
A random domain swapping method coupled with domain shuffling was developed by employing high-throughput construction and screening of chimeric endolysins. Yang et al. (Citation2015) attempted to develop an in vitro screening platform using a two-vector-based E. coli expression system. Through the in vitro screening platform, they selected a novel chimeric endolysin, named ClyR, forming a large, clear lysis zone in the Streptococcus dysgalactiae lawn. ClyR showed a broader host spectrum and lytic efficacy against Streptococcus spp. in pasteurized milk. Different research groups have developed recombinant endolysin targeting S. aureus using a similar screening strategy with some modifications. Son et al. (Citation2021) developed a rapid screening system employing SPN1S_lysRz protein, a spanin from Salmonella phage SPN1S, to lyse E. coli cells expressing a random domain swapping library of four different staphylococcal endolysins. They selected and investigated the lytic efficiency of the chimeric Lys109 endolysin. Lys109 exhibited highly improved lytic activity in killing S. aureus from milk and the surface of stainless steel. Further recombination was conducted by Lee, Kim, et al. (Citation2021). They constructed a library of chimeric endolysins by shuffling the domains of 12 natural staphylococcal endolysins. The most effective chimeric endolysin, ClyC, maintained its robust antibacterial activity in the Tris buffer (20 mM Tris-HCl, pH 8.0), milk, and animal blood and exhibited the protective capability of ClyC against MRSA-induced bacteremia in an in vivo mouse model.
Engineering of endolysins for biocontrol of gram-negative bacteria
The primary purpose of endolysin engineering for targeting Gram-negative bacteria is to penetrate the outer membrane because cell lysis of Gram-negative bacteria through external endolysin treatment is restricted due to the presence of an impermeable outer membrane (Oliveira et al. Citation2012). Although several natural endolysins have been reported to possess intrinsic OM permeability (Lai et al. Citation2011; Lim et al. Citation2014), the number of effective endolysins against Gram-negative bacteria is extremely low, and lytic activity is weak because the extent of penetration is limited. Previous studies have shown the effect of endolysins targeting Gram-negative bacteria in combination with physical (including heat and hydrostatic pressure) or chemical (including chloroform, EDTA, and organic acid) treatment (Endersen et al. Citation2015; Nakimbugwe et al. Citation2006; Oliveira et al. Citation2014a), but these results have limitations of safety issues and practical applications. In this section, we introduce four types of engineering strategies to target Gram-negative bacteria: (i) fusion protein with OM-penetrating peptides, termed “Artilysins”; (ii) fusion protein with OM-translocating domains derived from bacteria, termed “Lysocins,” fusion protein with receptor binding domains, coined “Innolysins”; and (iii) encapsulation of endolysin into liposome.
“Artilysin” strategy – fusion with OM-penetrating peptides
In the engineering of artilysins, peptides with diverse physicochemical properties (cationic, hydrophobic, or amphipathic) can be used to destabilize the negatively charged lipopolysaccharides (LPS) at the outer membrane (OM). Artilysins are chimeric endolysins combined with Gram-negative lysins and an OM-destabilizing peptide, which was first termed by Briers et al. (Citation2014). Art-175 is an example of artilysin, a fusion protein of a modified variant of endolysin KZ144 with SMAP-29 at the N-terminus end. Art-175 exhibited bactericidal activity against various Gram-negative strains, including Acinetobacter baumannii, Pseudomonas aeruginosa, and E. coli, and even resulted in the total eradication of large inocula of cells (≥108 CFU/mL) (Briers et al. Citation2014).
“Lysocin” & “innolysin” strategy – fusion with OM-translocating or receptor binding domains
Gram-negative endolysins may be engineered by exploiting colicin-like bacteriocins, which function in translocating the OM of bacteria and in delivery of endolysins to the peptidoglycan area (Heselpoth et al. Citation2019; Lukacik et al. Citation2012; Yan et al. Citation2017). Lysocins are fusion proteins that comprise part of the endolysin and bacteriocin responsible for transport in the outer membrane. Moreover, the fusion of phage RBPs into endolysins, coined as Innolysin, has recently been introduced as a novel approach to target Gram-negative bacteria. Zampara et al. (Citation2020) demonstrated that Innolysin Ec21 displayed potential bactericidal activity, causing more than 3 log reductions of E. coli cells resistant to third generation cephalosporins.
Formulation design for gram-negative endolysins
In addition to the fusion engineering approach, a formulation strategy for Gram-negative endolysins has been developed to enhance its permeability. Bai et al. (Citation2019) encapsulated the Salmonella endolysin BSP16Lys into cationic liposomes. The encapsulated BSPLys showed increased antibacterial activity up to 2.2 log and 1.6 log CFU/mL reductions in Salmonella Typhimurium and E. coli cells, respectively. Abouhmad et al. (Citation2017) conjugated a T4 lysozyme to cellulose nanocrystals (CNC), and this immobilization strategy significantly increased the bactericidal activity against E. coli and Pseudomonas mendocina compared to the free enzyme. The other encapsulating strategy utilizing non-biohazardous and biocompatible particles such as chitosan can also be applied to design Gram-negative endolysins for food safety because the pneumocidal activity of endolysin Cpl-1 is maintained in chitosan nanoparticles (Gondil et al. Citation2020). The summarized engineering strategies and examples of Gram-negative endolysins are listed in .
Challenges for the application of engineered phages and endolysins in the food industry
Given the short history of phage and endolysin applications after the emergence of antibiotic resistance (Altamirano and Barr Citation2019), improvements in engineering methodologies to design novel and optimized phages and endolysins are remarkable. However, we still encounter some basic and application issues that need to be refined for use as food antimicrobial agents. Before applying engineered phages and endolysins in the food industry, we should carefully consider their safety, stability, and production costs. Therefore, spurred on by huge food industry needs, continued research in related challenges is required to address the remaining concerns still seen in phage and phage-derived enzymes ().
Figure 2. Future research needs to address the remaining concerns still seen in the application of phages and endolysins in the food industry. The figure was created with Biorender (http://biorender.com).
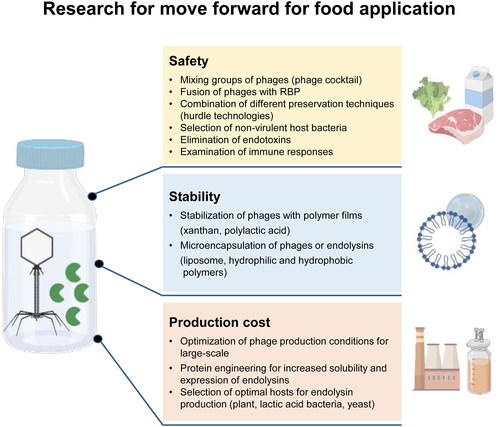
Safety
Safety requirements to be met by bacteriophages as therapeutics or food preservatives are: not to emerge phage resistance, not to contain virulence genes and other contaminants, and not to exhibit allergic and other immune reactions in humans. Bacteria have the natural ability to develop resistance to phages following repeated exposure to phage infections. Bacterial resistance against phages can be overcome by simply using an entirely different phage targeting the pathogen of interest, by mixing groups of phages (cocktail strategy), or by fuzing with RBP (Lenneman et al. Citation2021). In addition, hurdle technologies, which are a combination of different preservation techniques to eliminate or control pathogens, have been widely used to deal with the phage resistance problem in foodborne pathogens (Leistner Citation2000). In particular, synergistic treatments with chemicals, antibiotics, enzymes, and other antimicrobial peptides have shown promising results in decreasing phage resistance in bacteria (Becker, Foster-Frey, and Donovan Citation2008; Chang et al. Citation2017b; Chibeu et al. Citation2013; Djurkovic, Loeffler, and Fischetti Citation2005; García et al. Citation2010a). This combinational treatment can not only contribute to reducing the use of each agent but also decrease the likelihood of phage resistance.
Another safety issue, transfer of virulence factors and other contaminants originating from the bacterial host used for propagation, must be carefully considered before using phages as a bio-preservative. This indicates the need to select a virulent phage to reduce the possibility of transmission of virulence factors (Fernández et al. Citation2019). In addition, we should avoid selecting virulent drug-resistant pathogens as hosts for phage propagation (García et al. Citation2019). Nonetheless, if it is difficult to obtain non-virulent strains for large-scale phage production, gene manipulation should be attempted to construct non-virulent strains for safe phage production. Considering the large-scale phage production with host bacterium, it is a challenge to eliminate debris, bacterial contaminants, and live cells in crude phage lysate (Santos et al. Citation2010). Especially, endotoxins, which are potential contaminants from Gram-negative bacteria, should be removed during the phage purification process because of their side effects on human health. Thus, adequate physical and/or chemical purification should be adopted to produce toxin-free phage preparations. Furthermore, the cytotoxicity evaluation of purified recombinant phages should be included in the manufacturing process to ensure the safety of phage products.
The last hurdle for the use of engineered phages as food additives is the lack of enough clinical studies to provide a scientific basis for approval. Considering the fact that the engineered phages can influence human immunity via cellular uptake and recognition by the host (Popescu et al. Citation2021), it is critical to perform pre-clinical and clinical research associated with the interaction between the engineered phages and host immune cells to validate that the engineered phages are highly safe and harmless to human as a food additive. Although there have been several successful cases where natural phage has been applied as a therapeutic agent in the USA and Western Europe (Aslam et al. Citation2020; Wienhold, Lienau, and Witzenrath Citation2019), in the case of engineered phage, it has only been applied once to prevent Mycobacterium abscessus infection (Dedrick et al. Citation2019) and has not been approved for food application. In the first approval case of natural phage as a food additive by the FDA (FDA Citation2006), a number of safety studies of these Listeria-specific phages have provided the scientific basis for the first approval. Many phage-based products have been approved as food additives after the establishment of this first legal guideline. However, legal guidelines or regulations associated with application of engineered phages have not yet been established. Therefore, further studies of engineered phages related to their safety in humans will be helpful to overcome the conservative views toward the use of engineered phages and to establish the legal base for the approval of recombinant phages.
Stability
The second challenge is the stability of phages and endolysins in the food matrix to which they are applied. Engineered phages and endolysins have demonstrated strong antimicrobial performance on a laboratory scale. However, there may be a large discrepancy in their antimicrobial performance in food settings compared to in vitro settings because food is a giant complex of biometrics that have unique parameters, such as ionic composition, pH, salt concentration, temperature, and physical structure (Shannon, Radford, and Balamurugan Citation2020). The antimicrobial effects of phages could decrease due to particular intrinsic components in foods, such as whey proteins in bovine milk, protein, and fat in meat products (Ahmadi Citation2017; Gill et al. Citation2006). In the case of endolysin, the extent of cell lysis varies depending on the food conditions, such as pH, salt, temperature, and food matrix. Therefore, it is imperative to understand the optimal conditions for each endolysin prior to its application in foods. Further research is needed to determine how each food matrix plays a role in the antimicrobial activity of phages or endolysins and how these interactions are affected.
Several ways to apply phages and endolysins in a more stable form in food include using stabilizing gels and liposomes. Radford et al. (Citation2017) designed a novel packaging strategy with Listeria phage A511 and stabilizing gel coatings composed of a neutral polymer such as xanthan and polylactic acid (PLA). They demonstrated that when pre-cooked turkey breast was packaged in phage-coated PLA films, it significantly inhibited the growth of S. Typhimurium and L. monocytogenes for over 30 days. The microencapsulation of phages with sodium alginate prolonged the stability of phages targeting E. coli O157:H7 forming a stabilizing gel (Yin et al. Citation2021). In addition, for endolysins, studies on the structure of individual domains, the placement and characteristics of linker regions, charge distribution, cofactor usage, and affinity can provide insights for analyzing the effects of individual endolysins in specific foods (Shannon, Radford, and Balamurugan Citation2020). Therefore, advanced bioinformatics and structural analysis studies should be based to promote a more rational design of phages and endolysins with optimized antibacterial activity in certain food types.
Production cost
Another critical issue requiring further consideration is the low-cost, rapid, and easy production of phages and endolysins at a large-scale industrial level. The protocols for phage production in laboratories are well established, but data obtained in a laboratory are not always suitable for large-scale phage production (Kwok Citation2010). Several theoretical models have been proposed for large-scale phage production by considering kinetic parameters, such as bacterial-specific growth rate, burst size, adsorption rate, latency period, and eclipse time, and many experiments have obtained high concentrations of phages (García et al. Citation2019). The unit cost of phage production was estimated by Krysiak-Baltyn, Martin, and Gras (Citation2018), and Torress-Acosta et al. (2019, 2020). These studies suggest that production costs vary depending on the process conditions, including culture procedures, precipitation, and biological parameters, including bacterial growth and phage infection. To minimize production costs, future studies should consider other parameters such as energy transfer, the unit cost of each material, types of bioreactors, aeration proportion, and air inflow into the bioreactor.
The current cost of endolysin production is expected to be high, which may be an integral barrier to the implementation of endolysins as alternative antimicrobial agents (Oliveira et al. Citation2012). The high cost is mainly due to the need for rational recombination engineering and adequate, efficient, and safe expression systems for endolysin expression and purification. Therefore, technical approaches to increase the solubility and expression level of target endolysins, as well as cost-effective expression systems, are necessary to make endolysins commercially appealing food antimicrobial agents to consider investments.
There are many opportunities to reduce production costs and increase safety, ranging from the start line of protein expression to the downstream processing steps. After engineering and recombining endolysins in a specific expression system, downstream processing steps, such as cell harvesting, cell lysis, and purification, are conducted. In particular, in the case of protein expression in bacteria, endotoxin must be removed to avoid toxic effects. In addition, selecting optimal expression hosts (e.g., bacteria, yeast, or plants) early in the production process is an essential option for overexpression and low expression cost. Plants have been proposed as attractive hosts for producing recombinant proteins because the production costs are low. Also, proteins produced by plants are likely to be safer than proteins produced by animal cells or bacteria (Magnusdottir et al. Citation2013). According to Wittmann et al. (Citation2016), transgenic tomatoes expressing the Clavibacter michiganensis endolysin CMP1 did not show disease symptoms even though bacteria were not completely eliminated in tomatoes. Given the safety issues regarding the consumption of transgenic food products, much work remains to be done in plant expression and the production of endolysins.
In addition, secretion or release of specific endolysins by starter organisms or lactic acid bacteria (LAB) in fermentation processes is another good option for low treatment costs. LAB include the genera Lactococcus, Streptococcus, Lactobacillus, and Enterococcus and are generally recognized as safe (GRAS). Gaeng et al. (Citation2000) validated that the expression of L. monocytogenes endolysin Ply511 in Lactobacillus spp. is effective in fermented dairy products and has potential as a bio-preservative in foods. In another study, Gervasi et al. (Citation2014a, Citation2014b) expressed Clostridium perfringens amidase endolysin CP25L in the Lactobacillus johnsonii strain isolated from poultry. Through expression and secretion in this LAB, more than 2.6 log CFU/mL reduction of C. perfringens cells was observed in co-culture. Another study adopted Saccharomyces cerevisiae as a novel expression and surface-display platform for staphylococcal endolysin LysSA11. Chun, Bai, and Ryu (Citation2020) revealed that direct treatment of LysSA11-displaying yeast cells could reduce more than a 5-log of viable S. aureus cells within 3 h.
Concluding remarks: food safety perspective
From farm to fork, foods might be contaminated at any stage by various foodborne pathogens (Riggio et al. Citation2019). In general, the prevention and eradication of these foodborne pathogens have been conducted using various natural or chemical antimicrobial agents. Natural antimicrobial agents (e.g., organic acids, chitosan, plant extracts, essential oils, and bacteriocins) tend to possess weak and limited antimicrobial activity with a possible change in food texture and quality (Crozier-Dodson, Carter, and Zheng Citation2004; Juneja, Dwivedi, and Yan Citation2012). In addition, chemical antimicrobial agents in the food industry have raised consumer concern due to their known side effects (Pawlowska et al. Citation2012). Therefore, the demand for alternative antimicrobials that are safe for human consumption and not influential on the taste, texture, and nutritional quality of perishable food products, has directed attention to natural antimicrobial approaches. Furthermore, given the global antibiotic resistance crisis and the increase in MDR bacteria, the call to develop alternative methods that possess a different mechanism of action has intensified. Accordingly, many research groups have focused on phages and endolysins as potential candidates for controlling harmful microorganisms.
Phages are potentially evolving biological entities in that they can adapt themselves to evade changes; they are ubiquitous with an enormous number (about 1031), and they show high diversity. They provide an extensive genetic repository with a variety of viral sequences. According to Dion, Oechslin, and Moineau (Citation2020), the number of complete phage genomes in the NCBI nucleotide database site was 8,437, and the number of sequenced phage genomes doubled to 17,630 as of May 2021 (Ryu Citation2021). This is due to the recent advances in next-generation sequencing (NGS) technologies and long-range polymerases that are inexpensive and exhibit high fidelity. The more phage genome sequenced, the better insights into phage biology, including novel viral protein, phage resistance, and phage-host interaction, can be obtained in research communities. Fortunately, more than half of phage genes have been known to share low or no homology with other genes in the databases, suggesting that the novel viral genes studied up to now are just the tip of the iceberg (Ryu Citation2021). Considering its vast genetic sources, phage-derived antibacterial agents may possess more room for design than chemical antibiotics, allowing for continuous improvements in biologics. Along with this trend, the revolution in genetic technology and molecular engineering techniques will further fuel improvements in engineering methods for phages and endolysins in the future.
The potential for bioengineering of phages and endolysins is seemingly endless, including changing the lytic spectrum, optimizing the catalytic abilities, decreasing the likelihood of resistant bacteria, preventing the biofilms of pathogens, and improving its permeability to the outer membranes of Gram-negative bacteria. Many studies have introduced novel engineering strategies that employ phage and endolysin as control agents against Gram-negative pathogens. However, in the future, increased efforts are required for scientists, industries, and policymakers to maximize the bactericidal potential of phages and endolysins. Scientists should evaluate its efficacy under more realistic conditions, such as various fresh and/or processed foods and food production facilities, and evaluate its safety through pre-clinical and clinical studies to increase consumer acceptance. The industry needs continuous investments to adopt new techniques faster, and policymakers require to approve phage- and endolysin-based products when a solid scientific basis has been established. Positive public opinion will eventually bring them to the market if these efforts and considerations are made, and publicized. Now is the time to lay the technical foundation for developing phages and endolysins as food antimicrobial agents that efficiently control various foodborne pathogens and even MDR bacteria in the field of food safety.
Declaration of competing interest
All authors declare that there is no conflict of interest to publish the content of this article.
Additional information
Funding
References
- Abdelsattar, A. S., A. Dawooud, N. Rezk, S. Makky, A. Safwat, P. J. Richards, and A. El-Shibiny. 2021. How to train your phage: The recent efforts in phage training. Biologics 1 (2):70–88. doi: 10.3390/biologics1020005.
- Abedon, S. T., K. M. Danis-Wlodarczyk, D. J. Wozniak, and M. B. Sullivan. 2021. Improving phage-biofilm in vitro experimentation. Viruses 13 (6):1175. doi: 10.3390/v13061175.
- Abouhmad, A., T. Dishisha, M. A. Amin, and R. Hatti-Kaul. 2017. Immobilization to positively charged cellulose nanocrystals enhances the antibacterial activity and stability of hen egg white and T4 lysozyme. Biomacromolecules 18 (5):1600–8. doi: 10.1021/acs.biomac.7b00219.
- Abuladze, T., M. Li, M. Y. Menetrez, T. Dean, A. Senecal, and A. Sulakvelidze. 2008. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157: H7. Applied and Environmental Microbiology 74 (20):6230–8. doi: 10.1128/AEM.01465-08.
- Ahmadi, H. 2017. Thermal stability of encapsulated listeria bacteriophage and its efficacy against Listeria monocytogenes in ready-to-eat meats. PhD thesis., University of Guelph.
- Altamirano, F. L. G, and J. J. Barr. 2019. Phage therapy in the postantibiotic era. Clinical Microbiology Reviews 32 (2):e00066–18. doi: 10.1128/CMR.00066-18.
- Ando, H., S. Lemire, D. P. Pires, and T. K. Lu. 2015. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Systems 1 (3):187–96. doi: 10.1016/j.cels.2015.08.013.
- Aslam, S., E. Lampley, D. Wooten, M. Karris, C. Benson, S. Strathdee, and R. T. Schooley. 2020. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infectious Diseases 7 (9) doi: 10.1093/ofid/ofaa389.
- Au, A., H. Lee, T. Ye, U. Dave, and A. Rahman. 2021. Bacteriophages: Combating antimicrobial resistance in food-borne bacteria prevalent in agriculture. Microorganisms 10 (1):46. doi: 10.3390/microorganisms10010046.
- Azeredo, J, and I. W. Sutherland. 2008. The use of phages for the removal of infectious biofilms. Current Pharmaceutical Biotechnology 9 (4):261–6. doi: 10.2174/138920108785161604.
- Bai, J., E. Yang, P.-S. Chang, and S. Ryu. 2019. Preparation and characterization of endolysin-containing liposomes and evaluation of their antimicrobial activities against gram-negative bacteria. Enzyme and Microbial Technology 128:40–8. doi: 10.1016/j.enzmictec.2019.05.006.
- Bantawa, K., K. Rai, D. S. Limbu, and H. Khanal. 2018. Food-borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC Research Notes 11 (1):1–5. doi: 10.1186/s13104-018-3722-x.
- Bari, S. N., F. C. Walker, K. Cater, B. Aslan, and A. Hatoum-Aslan. 2017. Strategies for editing virulent staphylococcal phages using CRISPR-Cas10. ACS Synthetic Biology 6 (12):2316–25. doi: 10.1021/acssynbio.7b00240.
- Becker, S. C., J. Foster-Frey, A. J. Stodola, D. Anacker, and D. M. Donovan. 2009. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 443 (1–2):32–41. doi: 10.1016/j.gene.2009.04.023.
- Becker, S. C., J. Foster-Frey, and D. M. Donovan. 2008. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiology Letters 287 (2):185–91. doi: 10.1111/j.1574-6968.2008.01308.x.
- Born, Y., L. Fieseler, V. Thöny, N. Leimer, B. Duffy, and M. J. Loessner. 2017. Engineering of bacteriophages Y2:: DpoL1-C and Y2:: luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Applied and Environmental Microbiology 83 (12):e00341–00317. doi: 10.1128/AEM.00341-17.
- Borysowski, J., B. Weber-Dabrowska, and A. Górski. 2006. Bacteriophage endolysins as a novel class of antibacterial agents. Experimental Biology and Medicine (Maywood, N.J.) 231 (4):366–77. doi: 10.1177/153537020623100402.
- Bren, L. 2007. Bacteria-eating virus approved as food additive. FDA Consumer 41 (1):20–2.
- Briers, Y., M. Walmagh, V. Van Puyenbroeck, A. Cornelissen, W. Cenens, A. Aertsen, H. Oliveira, J. Azeredo, G. Verween, J.-P. Pirnay, et al. 2014. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5 (4):e01379–01314. doi: 10.1128/mBio.01379-14.
- Brüssow, H, and R. W. Hendrix. 2002. Phage genomics: Small is beautiful. Cell 108 (1):13–6. doi: 10.1016/S0092-8674(01)00637-7.
- Bundy, B. C., M. J. Franciszkowicz, and J. R. Swartz. 2008. Escherichia coli-based cell-free synthesis of virus-like particles. Biotechnology and Bioengineering 100 (1):28–37. doi: 10.1002/bit.21716.
- Carlton, R., W. Noordman, B. Biswas, E. De Meester, and M. J. Loessner. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regulatory Toxicology and Pharmacology : RTP 43 (3):301–12. doi: 10.1016/j.yrtph.2005.08.005.
- CDC. 2019. Antibiotic resistance threats in the United States, 2019. In: 2019 AR Threats Report. Center for Disease Control and Prevention.
- CDC. 2020. About antibiotic resistance. In: Public Health Reports. Center for Disease Control and Prevention.
- CDC. 2021. Listeria Outbreak Linked to Queso Fresco Made by El Abuelito Cheese Inc. Center for Disease Control and Prevention.
- Cha, Y., J. Chun, B. Son, and S. Ryu. 2019. Characterization and genome analysis of Staphylococcus aureus podovirus CSA13 and its anti-biofilm capacity. Viruses 11 (1):54. doi: 10.3390/v11010054.
- Chang, Y., H. Yoon, D.-H. Kang, P.-S. Chang, and S. Ryu. 2017b. Endolysin LysSA97 is synergistic with carvacrol in controlling Staphylococcus aureus in foods. International Journal of Food Microbiology 244:19–26. doi: 10.1016/j.ijfoodmicro.2016.12.007.
- Chang, Y., M. Kim, and S. Ryu. 2017a. Characterization of a novel endolysin LysSA11 and its utility as a potent biocontrol agent against Staphylococcus aureus on food and utensils. Food Microbiology 68:112–20. doi: 10.1016/j.fm.2017.07.004.
- Channabasappa, S., R. Chikkamadaiah, M. Durgaiah, S. Kumar, K. Ramesh, A. Sreekanthan, and B. Sriram. 2018. Efficacy of chimeric ectolysin P128 in drug-resistant Staphylococcus aureus bacteraemia in mice. The Journal of Antimicrobial Chemotherapy 73 (12):3398–404. doi: 10.1093/jac/dky365.
- Chen, Y., H. Batra, J. Dong, C. Chen, V. B. Rao, and P. Tao. 2019. Genetic engineering of bacteriophages against infectious diseases. Frontiers in Microbiology 10:954. doi: 10.3389/fmicb.2019.00954.
- Cheng, Q, and V. A. Fischetti. 2007. Mutagenesis of a bacteriophage lytic enzyme PlyGBS significantly increases its antibacterial activity against group B streptococci. Applied Microbiology and Biotechnology 74 (6):1284–91. doi: 10.1007/s00253-006-0771-1.
- Chibeu, A., L. Agius, A. Gao, P. M. Sabour, A. M. Kropinski, and S. Balamurugan. 2013. Efficacy of bacteriophage LISTEX™ P100 combined with chemical antimicrobials in reducing Listeria monocytogenes in cooked turkey and roast beef. International Journal of Food Microbiology 167 (2):208–14. doi: 10.1016/j.ijfoodmicro.2013.08.018.
- Chun, J., J. Bai, and S. Ryu. 2020. Yeast surface display system for facilitated production and application of phage endolysin. ACS Synthetic Biology 9 (3):508–16. doi: 10.1021/acssynbio.9b00360.
- Cieplak, T., N. Soffer, A. Sulakvelidze, and D. S. Nielsen. 2018. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 9 (5):391–9.
- Crozier-Dodson, B. A., M. Carter, and Z. Zheng. 2004. Formulating food safety: An overview of antimicrobial ingredients. Food Safety Magazine.
- Dedrick, R., C. Guerrero-Bustamante, R. Garlena, D. Russell, K. Ford, K. Harris, K. Gilmour, J. Soothill, D. Jacobs-Sera, R. Schooley, et al. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature Medicine 25 (5):730–3. doi: 10.1038/s41591-019-0437-z.
- DeVita, M. D., R. K. Wadhera, M. L. Theis, and S. C. Ingham. 2007. Assessing the potential of Streptococcus pyogenes and Staphylococcus aureus transfer to foods and customers via a survey of hands, hand‐contact surfaces and food‐contact surfaces at foodservice facilities. Journal of Foodservice 18 (2):76–9. doi: 10.1111/j.1745-4506.2007.00049.x.
- Díez-Martínez, R., H. D. De Paz, E. García-Fernández, N. Bustamante, C. W. Euler, V. A. Fischetti, M. Menendez, and P. García. 2015. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. The Journal of Antimicrobial Chemotherapy 70 (6):1763–73. doi: 10.1093/jac/dkv038.
- Díez-Martínez, R., H. de Paz, H. de Paz, N. Bustamante, E. García, M. Menéndez, and P. García. 2013. Improving the lethal effect of Cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrobial Agents and Chemotherapy 57 (11):5355–65.
- Dion, M. B., F. Oechslin, and S. Moineau. 2020. Phage diversity, genomics and phylogeny. Nature Reviews Microbiology 18 (3):125–38.
- Djurkovic, S., J. M. Loeffler, and V. A. Fischetti. 2005. Synergistic killing of Streptococcus pneumoniae with the bacteriophage lytic enzyme Cpl-1 and penicillin or gentamicin depends on the level of penicillin resistance. Antimicrobial Agents and Chemotherapy 49 (3):1225–8. doi: 10.1128/AAC.49.3.1225-1228.2005.
- Dong, J., C. Chen, Y. Liu, J. Zhu, M. Li, V. B. Rao, and P. Tao. 2021. Engineering T4 bacteriophage for in vivo display by type V CRISPR-cas genome editing. ACS Synthetic Biology 10 (10):2639–48. doi: 10.1021/acssynbio.1c00251.
- Dublanchet, A, and S. Bourne. 2007. The epic of phage therapy. The Canadian Journal of Infectious Diseases & Medical Microbiology = Journal Canadien Des Maladies Infectieuses et de la Microbiologie Medicale 18 (1):15–8. doi: 10.1155/2007/365761.
- Duong, M. M., C. M. Carmody, Q. Ma, J. E. Peters, and S. R. Nugen. 2020. Optimization of T4 phage engineering via CRISPR/Cas9. Scientific Reports 10 (1):1–9. doi: 10.1038/s41598-020-75426-6.
- Endersen, L., C. Guinane, C. Johnston, H. Neve, A. Coffey, P. Ross, O. McAuliffe, and J. O’Mahony. 2015. Genome analysis of Cronobacter phage vB_CsaP_Ss1 reveals an endolysin with potential for biocontrol of Gram-negative bacterial pathogens. The Journal of General Virology 96 (Pt 2):463–77. doi: 10.1099/vir.0.068494-0.
- English, B. K, and A. H. Gaur. 2010. The use and abuse of antibiotics and the development of antibiotic resistance. In Hot topics in infection and immunity in children VI, eds. A. Finn, N. Curtis, and A. J. Pollard, 73–82. New York: Springer.
- Fan, J., Z. Zeng, K. Mai, Y. Yang, J. Feng, Y. Bai, B. Sun, Q. Xie, Y. Tong, and J. Ma. 2016. Preliminary treatment of bovine mastitis caused by Staphylococcus aureus, with trx-SA1, recombinant endolysin of S. aureus bacteriophage IME-SA1. Veterinary Microbiology 191:65–71.
- Favor, A. H., C. D. Llanos, M. D. Youngblut, and J. A. Bardales. 2020. Optimizing bacteriophage engineering through an accelerated evolution platform. Scientific Reports 10 (1):1–10. doi: 10.1038/s41598-020-70841-1.
- FDA. 2006. Department of health and Human services Food and Drug Administration, 21 CFR Part 172 [Docket No. 2002F–0316 (formerly 02F-0316)]. Food Additives Permitted for Direct Addition to Food for Human Consumption: 47729-47732.
- FDA. 2008. Guidance for industry: Guide to minimize microbial food safety hazards of fresh-cut fruits and vegetables. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/guidance-industry-guide-minimizemicrobial-food-safety-hazards-fresh-fruits-and-vegetables
- Fernández, L., D. Gutiérrez, P. García, and A. Rodríguez. 2019. The perfect bacteriophage for therapeutic applications—A quick guide. Antibiotics 8 (3):126. doi: 10.3390/antibiotics8030126.
- Fischetti, V. A. 2003. Novel method to control pathogenic bacteria on human mucous membranes. Annals of the New York Academy of Sciences 987:207–14. doi: 10.1111/j.1749-6632.2003.tb06050.x.
- Fischetti, V. A. 2005. Bacteriophage lytic enzymes: Novel anti-infectives. Trends in Microbiology 13 (10):491–6. doi: 10.1016/j.tim.2005.08.007.
- Francino, M. 2016. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Frontiers in Microbiology 6:1543. doi: 10.3389/fmicb.2015.01543.
- Fruciano, D. E, and S. Bourne. 2007. Phage as an antimicrobial agent: d’Herelle’s heretical theories and their role in the decline of phage prophylaxis in the West. The Canadian Journal of Infectious Diseases & Medical Microbiology = Journal Canadien Des Maladies Infectieuses et de la Microbiologie Medicale 18 (1):19–26. doi: 10.1155/2007/976850.
- Gaeng, S., S. Scherer, H. Neve, and M. J. Loessner. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Applied and Environmental Microbiology 66 (7):2951–8. doi: 10.1128/AEM.66.7.2951-2958.2000.
- Garamella, J., R. Marshall, M. Rustad, and V. Noireaux. 2016. The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synthetic Biology 5 (4):344–55. doi: 10.1021/acssynbio.5b00296.
- García, P., B. Martínez, L. Rodríguez, and A. Rodríguez. 2010a. Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. International Journal of Food Microbiology 141 (3):151–5. doi: 10.1016/j.ijfoodmicro.2010.04.029.
- García, P., L. Rodríguez, A. Rodríguez, and B. Martínez. 2010b. Food biopreservation: Promising strategies using bacteriocins, bacteriophages and endolysins. Trends in Food Science & Technology 21 (8):373–82. doi: 10.1016/j.tifs.2010.04.010.
- García, R., S. Latz, J. Romero, G. Higuera, K. García, and R. Bastías. 2019. Bacteriophage production models: An overview. Frontiers in Microbiology 10:1187. doi: 10.3389/fmicb.2019.01187.
- Garenne, D., S. Thompson, A. Brisson, A. Khakimzhan, and V. Noireaux. 2021. The all-E. coliTXTL toolbox 3.0: New capabilities of a cell-free synthetic biology platform. Synthetic Biology (Oxford, England) 6 (1):ysab017. 2021doi: 10.1093/synbio/ysab017.
- Gerstmans, H., B. Criel, and Y. Briers. 2018. Synthetic biology of modular endolysins. Biotechnology Advances 36 (3):624–40. doi: 10.1016/j.biotechadv.2017.12.009.
- Gervasi, T., N. Horn, U. Wegmann, G. Dugo, A. Narbad, and M. J. Mayer. 2014a. Expression and delivery of an endolysin to combat Clostridium perfringens. Applied Microbiology and Biotechnology 98 (6):2495–505. doi: 10.1007/s00253-013-5128-y.
- Gervasi, T., R. Lo Curto, E. Minniti, A. Narbad, and M. J. Mayer. 2014b. Application of Lactobacillus johnsonii expressing phage endolysin for control of Clostridium perfringens. Letters in Applied Microbiology 59 (4):355–61. doi: 10.1111/lam.12298.
- Gharieb, R. M. A., M. F. Saad, A. S. Mohamed, and Y. H. Tartor. 2020. Characterization of two novel lytic bacteriophages for reducing biofilms of zoonotic multidrug-resistant Staphylococcus aureus and controlling their growth in milk. LWT 124:109145. doi: 10.1016/j.lwt.2020.109145.
- Gibson, D. G., L. Young, R.-Y. Chuang, J. C. Venter, C. A. Hutchison, and H. O. Smith. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6 (5):343–5. doi: 10.1038/nmeth.1318.
- Gill, J., P. Sabour, K. Leslie, and M. Griffiths. 2006. Bovine whey proteins inhibit the interaction of Staphylococcus aureus and bacteriophage K. Journal of Applied Microbiology 101 (2):377–86. doi: 10.1111/j.1365-2672.2006.02918.x.
- Gondil, V. S., T. Dube, J. J. Panda, R. M. Yennamalli, K. Harjai, and S. Chhibber. 2020. Comprehensive evaluation of chitosan nanoparticle based phage lysin delivery system; a novel approach to counter S. pneumoniae infections. International Journal of Pharmaceutics 573:118850. doi: 10.1016/j.ijpharm.2019.118850.
- Goodridge, L. D, and B. Bisha. 2011. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 1 (3):130–7. doi: 10.4161/bact.1.3.17629.
- Guenther, S, and M. J. Loessner. 2011. Bacteriophage biocontrol of Listeria monocytogenes on soft ripened white mold and red-smear cheeses. Bacteriophage 1 (2):94–100. doi: 10.4161/bact.1.2.15662.
- Guo, M., C. Feng, J. Ren, X. Zhuang, Y. Zhang, Y. Zhu, K. Dong, P. He, X. Guo, and J. Qin. 2017. A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa. Frontiers in Microbiology 8 (293):293.
- Gutiérrez, D., L. Fernández, B. Martínez, P. Ruas-Madiedo, P. García, and A. Rodríguez. 2017. Real-time assessment of Staphylococcus aureus biofilm disruption by phage-derived proteins. Frontiers in Microbiology 8:1632. doi: 10.3389/fmicb.2017.01632.
- Gutiérrez, D., P. Ruas-Madiedo, B. Martínez, A. Rodríguez, and P. García. 2014. Effective removal of staphylococcal biofilms by the endolysin LysH5. PloS One 9 (9):e107307. doi: 10.1371/journal.pone.0107307.
- Gutiérrez, D., V. Garrido, L. Fernández, S. Portilla, A. Rodríguez, M. J. Grilló, and P. García. 2020. Phage lytic protein LysRODI prevents staphylococcal mastitis in mice. Frontiers in Microbiology 11:7. doi: 10.3389/fmicb.2020.00007.
- Hassan, A. Y., J. T. Lin, N. Ricker, and H. Anany. 2021. The age of phage: Friend or foe in the new dawn of therapeutic and biocontrol applications? Pharmaceuticals 14 (3):199. doi: 10.3390/ph14030199.
- Hatoum-Aslan, A. 2018. Phage genetic engineering using CRISPR–Cas systems. Viruses 10 (6):335. doi: 10.3390/v10060335.
- Heselpoth, R. D., C. W. Euler, R. Schuch, and V. A. Fischetti. 2019. Lysocins: Bioengineered antimicrobials that deliver lysins across the outer membrane of gram-negative bacteria. Antimicrobial Agents and Chemotherapy 63 (6):e00342–00319. doi: 10.1128/AAC.00342-19.
- Horgan, M., G. O’Flynn, J. Garry, J. Cooney, A. Coffey, G. Fitzgerald, P. Ross, and O. McAuliffe. 2009. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Applied and Environmental Microbiology 75 (3):872–4. doi: 10.1128/AEM.01831-08.
- Huss, P, and S. Raman. 2020. Engineered bacteriophages as programmable biocontrol agents. Current Opinion in Biotechnology 61:116–21. doi: 10.1016/j.copbio.2019.11.013.
- Ibarra-Sánchez, L. A., M. L. Van Tassell, and M. J. Miller. 2018. Antimicrobial behavior of phage endolysin PlyP100 and its synergy with nisin to control Listeria monocytogenes in Queso Fresco. Food Microbiology 72:128–34. doi: 10.1016/j.fm.2017.11.013.
- Jaschke, P. R., E. K. Lieberman, J. Rodriguez, A. Sierra, and D. Endy. 2012. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology 434 (2):278–84. doi: 10.1016/j.virol.2012.09.020.
- Jensen, J., A. Parks, S. Adhya, A. Rattray, and D. Court. 2020. λ Recombineering used to engineer the genome of phage T7. Antibiotics 9 (11):805. doi: 10.3390/antibiotics9110805.
- Jewett, M. C, and J. R. Swartz. 2004. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnology and Bioengineering 86 (1):19–26. doi: 10.1002/bit.20026.
- Jończyk, E., M. Kłak, R. Międzybrodzki, and A. Górski. 2011. The influence of external factors on bacteriophages-review. Folia Microbiologica 56 (3):191–200. doi: 10.1007/s12223-011-0039-8.
- Juneja, V. K., H. P. Dwivedi, and X. Yan. 2012. Novel natural food antimicrobials. Annual Review of Food Science and Technology 3:381–403. doi: 10.1146/annurev-food-022811-101241.
- Junttila, J. R., S. Niemelä, and J. Hirn. 1988. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. The Journal of Applied Bacteriology 65 (4):321–7. doi: 10.1111/j.1365-2672.1988.tb01898.x.
- Jurczak-Kurek, A., T. Gąsior, B. Nejman-Faleńczyk, S. Bloch, A. Dydecka, G. Topka, A. Necel, M. Jakubowska-Deredas, M. Narajczyk, M. Richert, et al. 2016. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Scientific Reports 6:34338–17. doi: 10.1038/srep34338.
- Kasman, L. M, and L. D. Porter. 2020. Bacteriophages. StatPearls [Internet]
- Kilcher, S., P. Studer, C. Muessner, J. Klumpp, and M. J. Loessner. 2018. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proceedings of the National Academy of Sciences 115 (3):567–72. doi: 10.1073/pnas.1714658115.
- Kim, J., H. Park, J. Kim, J. H. Kim, J. I. Jung, S. Cho, S. Ryu, and B. Jeon. 2019. Comparative analysis of aerotolerance, antibiotic resistance, and virulence gene prevalence in Campylobacter jejuni isolates from retail raw chicken and duck meat in South Korea. Microorganisms 7 (10):433. doi: 10.3390/microorganisms7100433.
- Kim, M, and S. Ryu. 2012. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Molecular Microbiology 86 (2):411–25. doi: 10.1111/j.1365-2958.2012.08202.x.
- Kiro, R., D. Shitrit, and U. Qimron. 2014. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biology 11 (1):42–4. doi: 10.4161/rna.27766.
- Kong, M., H. Na, N.-C. Ha, and S. Ryu. 2019. LysPBC2, a novel endolysin harboring a Bacillus cereus spore binding domain. Applied and Environmental Microbiology 85 (5):e02462–02418. doi: 10.1128/AEM.02462-18.
- Krysiak-Baltyn, K., G. J. Martin, and S. L. Gras. 2018. Computational modelling of large scale phage production using a two-stage batch process. Pharmaceuticals 11 (2):31. doi: 10.3390/ph11020031.
- Kumar, G. D. S. Ravishankar, and V. Juneja. 2017. Microbial Control and Food Preservation. New York: Springer.
- Kutter, E, and A. Sulakvelidze. 2004. Bacteriophages: Biology and applications. Boca Raton, Florida: CRC press.
- Kwok, R. 2010. Five hard truths for synthetic biology. Nature 463 (7279):288–90. doi: 10.1038/463288a.
- Lai, M.-J., K.-C. Chang, S.-W. Huang, C.-H. Luo, P.-Y. Chiou, C.-C. Wu, and N.-T. Lin. 2016. The tail associated protein of Acinetobacter baumannii phage ΦAB6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PloS One 11 (4):e0153361. doi: 10.1371/journal.pone.0153361.
- Lai, M.-J., N.-T. Lin, A. Hu, P.-C. Soo, L.-K. Chen, L.-H. Chen, and K.-C. Chang. 2011. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Applied Microbiology and Biotechnology 90 (2):529–39. doi: 10.1007/s00253-011-3104-y.
- Latka, A., S. Lemire, D. Grimon, D. Dams, B. Maciejewska, T. Lu, Z. Drulis-Kawa, and Y. Briers. 2021. Engineering the modular receptor-binding proteins of Klebsiella phages switches their capsule serotype specificity. mBio 12 (3):e00455–00421. doi: 10.1128/mBio.00455-21.
- Le, S., X. He, Y. Tan, G. Huang, L. Zhang, R. Lux, W. Shi, and F. Hu. 2013. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PloS One 8 (7):e68562. doi: 10.1371/journal.pone.0068562.
- Lee, C., J. Kim, B. Son, and S. Ryu. 2021. Development of Advanced Chimeric Endolysin to Control Multidrug-Resistant Staphylococcus aureus through Domain Shuffling. ACS Infectious Diseases 7 (8):2081–92. 2021. doi: 10.1021/acsinfecdis.0c00812.
- Lee, Y., B. Son, Y. Cha, and S. Ryu. 2021. Characterization and Genomic Analysis of PALS2, a Novel Staphylococcus Jumbo Bacteriophage. Frontiers in Microbiology 12 (395) doi: 10.3389/fmicb.2021.622755.
- Leistner, L. 2000. Basic aspects of food preservation by hurdle technology. International Journal of Food Microbiology 55 (1-3):181–6. doi: 10.1016/s0168-1605(00)00161-6.
- Lenneman, B. R., J. Fernbach, M. J. Loessner, T. K. Lu, and S. Kilcher. 2021. Enhancing phage therapy through synthetic biology and genome engineering. Current Opinion in Biotechnology 68:151–9. doi: 10.1016/j.copbio.2020.11.003.
- León, M, and R. Bastías. 2015. Virulence reduction in bacteriophage resistant bacteria. Frontiers in Microbiology 6:343. doi: 10.3389/fmicb.2015.00343.
- Leverentz, B., W. S. Conway, M. J. Camp, W. J. Janisiewicz, T. Abuladze, M. Yang, R. Saftner, and A. Sulakvelidze. 2003. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Applied and Environmental Microbiology 69 (8):4519–26. doi: 10.1128/AEM.69.8.4519-4526.2003.
- Leverentz, B., W. S. Conway, W. Janisiewicz, and M. J. Camp. 2004. Optimizing concentration and timing of a phage spray application to reduce Listeria monocytogenes on honeydew melon tissue. Journal of Food Protection 67 (8):1682–6. doi: 10.4315/0362-028x-67.8.1682.
- Li, Y.-K., H. Chen, M. Shu, C. Zhong, Y. Bi, H.-H. Yang, and G.-P. Wu. 2021. Isolation, characterization and application of an alkaline resistant virulent bacteriophage JN01 against Escherichia coli O157: H7 in milk and beef. LWT 144:111266. doi: 10.1016/j.lwt.2021.111266.
- Lim, J.-A., H. Shin, S. Heu, and S. Ryu. 2014. Exogenous lytic activity of SPN9CC endolysin against gram-negative bacteria. Journal of Microbiology and Biotechnology 24 (6):803–11. doi: 10.4014/jmb.1403.03035.
- Lin, D. M., B. Koskella, and H. C. Lin. 2017. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World Journal of Gastrointestinal Pharmacology and Therapeutics 8 (3):162–73. doi: 10.4292/wjgpt.v8.i3.162.
- Lineback, D. R., A. Pirlet, J. W. Van Der Kamp, and R. Wood. 2009. Globalization, food safety issues & role of international standards. Quality Assurance and Safety of Crops & Foods 1 (1):23–7. doi: 10.1111/j.1757-837X.2009.00005.x.
- Liu, A., Y. Wang, X. Cai, S. Jiang, X. Cai, L. Shen, Y. Liu, G. Han, S. Chen, J. Wang, et al. 2019. Characterization of endolysins from bacteriophage LPST10 and evaluation of their potential for controlling Salmonella Typhimurium on lettuce. LWT 114:108372. doi: 10.1016/j.lwt.2019.108372.
- Loc-Carrillo, C, and S. T. Abedon. 2011. Pros and cons of phage therapy. Bacteriophage 1 (2):111–4. doi: 10.4161/bact.1.2.14590.
- Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science (New York, N.Y.) 294 (5549):2170–2. doi: 10.1126/science.1066869.
- Loessner, M. J. 2005. Bacteriophage endolysins-current state of research and applications. Current Opinion in Microbiology 8 (4):480–7. doi: 10.1016/j.mib.2005.06.002.
- Loessner, M. J., C. Rees, G. Stewart, and S. Scherer. 1996. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Applied and Environmental Microbiology 62 (4):1133–40. doi: 10.1128/aem.62.4.1133-1140.1996.
- López-Cuevas, O., J. Medrano-Félix, N. Castro-Del Campo, and C. Chaidez. 2019. Bacteriophage applications for fresh produce food safety. International Journal of Environmental Health Research 31 (6):687–702. doi: 10.1080/09603123.2019.1680819.
- Love, M. J., D. Coombes, S. H. Manners, G. S. Abeysekera, C. Billington, and R. C. Dobson. 2021. The molecular basis for Escherichia coli O157: H7 phage FAHEc1 endolysin function and protein engineering to increase thermal stability. Viruses 13 (6):1101. doi: 10.3390/v13061101.
- Low, L. Y., C. Yang, M. Perego, A. Osterman, and R. Liddington. 2011. Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins. Journal of Biological Chemistry 286 (39):34391–403. doi: 10.1074/jbc.M111.244160.
- Lu, T. K, and J. J. Collins. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences of the United States of America 104 (27):11197–202. doi: 10.1073/pnas.0704624104.
- Lukacik, P., T. Barnard, P. Keller, K. Chaturvedi, N. Seddiki, J. Fairman, N. Noinaj, T. Kirby, J. Henderson, A. Steven, et al. 2012. Structural engineering of a phage lysin that targets Gram-negative pathogens. Proceedings of the National Academy of Sciences of the United States of America 109 (25):9857–62. doi: 10.1073/pnas.1203472109.
- Magnusdottir, A., H. Vidarsson, J. M. Björnsson, and B. L. Örvar. 2013. Barley grains for the production of endotoxin-free growth factors. Trends in Biotechnology 31 (10):572–80. doi: 10.1016/j.tibtech.2013.06.002.
- Mahichi, F., A. J. Synnott, K. Yamamichi, T. Osada, and Y. Tanji. 2009. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiology Letters 295 (2):211–7. doi: 10.1111/j.1574-6968.2009.01588.x.
- Manyi-Loh, C., S. Mamphweli, E. Meyer, and A. Okoh. 2018. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 23 (4):795. doi: 10.3390/molecules23040795.
- Mao, J., M. Schmelcher, W. J. Harty, J. Foster-Frey, and D. M. Donovan. 2013. Chimeric Ply187 endolysin kills Staphylococcus aureus more effectively than the parental enzyme. FEMS Microbiology Letters 342 (1):30–6. doi: 10.1111/1574-6968.12104.
- Marinelli, L., M. Piuri, Z. Swigonová, A. Balachandran, L. Oldfield, J. van Kessel, and G. Hatfull. 2008. BRED: A simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PloS One 3 (12):e3957. doi: 10.1371/journal.pone.0003957.
- Martel, B, and S. Moineau. 2014. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Research 42 (14):9504–13. doi: 10.1093/nar/gku628.
- Matsuzaki, S., M. Rashel, J. Uchiyama, S. Sakurai, T. Ujihara, M. Kuroda, M. Ikeuchi, T. Tani, M. Fujieda, H. Wakiguchi, et al. 2005. Bacteriophage therapy: A revitalized therapy against bacterial infectious diseases. Journal of Infection and Chemotherapy : Official Journal of the Japan Society of Chemotherapy 11 (5):211–9. doi: 10.1007/s10156-005-0408-9.
- Mayer, M. J., J. Payne, M. J. Gasson, and A. Narbad. 2010. Genomic sequence and characterization of the virulent bacteriophage phiCTP1 from Clostridium tyrobutyricum and heterologous expression of its endolysin. Applied and Environmental Microbiology 76 (16):5415–22. doi: 10.1128/AEM.00989-10.
- Mayer, M. J., V. Garefalaki, R. Spoerl, A. Narbad, and R. Meijers. 2011. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. Journal of Bacteriology 193 (19):5477–86. doi: 10.1128/JB.00439-11.
- Meaden, S, and B. Koskella. 2013. Exploring the risks of phage application in the environment. Frontiers in Microbiology 4:358. doi: 10.3389/fmicb.2013.00358.
- Montgomery, H., S. A. Haughey, and C. T. Elliott. 2020. Recent food safety and fraud issues within the dairy supply chain (2015–2019). Global Food Security 26:100447.
- Moye, Z. D., J. Woolston, and A. Sulakvelidze. 2018. Bacteriophage applications for food production and processing. Viruses 10 (4):205. doi: 10.3390/v10040205.
- Nakimbugwe, D., B. Masschalck, M. Atanassova, A. Zewdie-Bosüner, and C. W. Michiels. 2006. Comparison of bactericidal activity of six lysozymes at atmospheric pressure and under high hydrostatic pressure. International Journal of Food Microbiology 108 (3):355–63. doi: 10.1016/j.ijfoodmicro.2005.11.021.
- Nelson, D. C., M. Schmelcher, L. Rodríguez-Rubio, J. Klumpp, D. G. Pritchard, S. Dong, and D. M. Donovan. 2012. Endolysins as antimicrobials. Advances in Virus Research 83:299–365.
- Obeso, J. M., B. Martínez, A. Rodríguez, and P. García. 2008. Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. International Journal of Food Microbiology 128 (2):212–8. doi: 10.1016/j.ijfoodmicro.2008.08.010.
- OECD. 2018. Stemming the superbug tide.
- Oliveira, H., J. Azeredo, R. Lavigne, and L. D. Kluskens. 2012. Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends in Food Science & Technology 28 (2):103–15. doi: 10.1016/j.tifs.2012.06.016.
- Oliveira, H., L. D. Melo, S. B. Santos, F. L. Nóbrega, E. C. Ferreira, N. Cerca, J. Azeredo, and L. D. Kluskens. 2013. Molecular aspects and comparative genomics of bacteriophage endolysins. Journal of Virology 87 (8):4558–70. doi: 10.1128/JVI.03277-12.
- Oliveira, H., V. Thiagarajan, M. Walmagh, S. Sillankorva, R. Lavigne, M. T. Neves-Petersen, L. D. Kluskens, and J. Azeredo. 2014a. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram-negative pathogens in presence of weak acids. PloS One 9 (10):e108376. doi: 10.1371/journal.pone.0108376.
- Oliveira, M., I. Viñas, P. Colàs, M. Anguera, J. Usall, and M. Abadias. 2014b. Effectiveness of a bacteriophage in reducing Listeria monocytogenes on fresh-cut fruits and fruit juices. Food Microbiology 38:137–42. doi: 10.1016/j.fm.2013.08.018.
- Oliver, S. P., B. M. Jayarao, and R. A. Almeida. 2005. Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implications. Foodborne Pathogens and Disease 2 (2):115–29. doi: 10.1089/fpd.2005.2.115.
- Pagan, F. 1981. Antibiotics for gram-positive organisms. British Journal of Hospital Medicine 25 (1):24–7.
- Park, H., J. Kim, M. Kim, Y. Park, and S. Ryu. 2021. Development of new strategy combining heat treatment and phage cocktail for post-contamination prevention. Food Research International (Ottawa, Ont.) 145:110415. doi: 10.1016/j.foodres.2021.110415.
- Pastagia, M., C. Euler, P. Chahales, J. Fuentes-Duculan, J. G. Krueger, and V. A. Fischetti. 2011. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains . Antimicrobial Agents and Chemotherapy 55 (2):738–44. doi: 10.1128/AAC.00890-10.
- Pastagia, M., R. Schuch, V. A. Fischetti, and D. B. Huang. 2013. Lysins: The arrival of pathogen-directed anti-infectives. Journal of Medical Microbiology 62 (Pt 10):1506–16. doi: 10.1099/jmm.0.061028-0.
- Pawlowska, A. M., E. Zannini, A. Coffey, and E. K. Arendt. 2012. “ Green preservatives”: Combating fungi in the food and feed industry by applying antifungal lactic acid bacteria. Advances in Food and Nutrition Research 66:217–38.
- Perera, M. N., T. Abuladze, M. Li, J. Woolston, and A. Sulakvelidze. 2015. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiology 52:42–8. doi: 10.1016/j.fm.2015.06.006.
- Pires, D. P., S. Cleto, S. Sillankorva, J. Azeredo, and T. K. Lu. 2016. Genetically engineered phages: A review of advances over the last decade. Microbiology and Molecular Biology Reviews : MMBR 80 (3):523–43. doi: 10.1128/MMBR.00069-15.
- Połaska, M, and B. Sokołowska. 2019. Bacteriophages-a new hope or a huge problem in the food industry. AIMS Microbiology 5 (4):324–46. doi: 10.3934/microbiol.2019.4.324.
- Popescu, M., J. D. Van Belleghem, A. Khosravi, and P. L. Bollyky. 2021. Bacteriophages and the immune system. Annual Review of Virology 8 (1):415–35. doi: 10.1146/annurev-virology-091919-074551.
- Radford, D., B. Guild, P. Strange, R. Ahmed, L.-T. Lim, and S. Balamurugan. 2017. Characterization of antimicrobial properties of Salmonella phage Felix O1 and Listeria phage A511 embedded in xanthan coatings on Poly (lactic acid) films. Food Microbiology 66:117–28. doi: 10.1016/j.fm.2017.04.015.
- Ramirez-Chamorro, L., P. Boulanger, and O. Rossier. 2021. Strategies for bacteriophage T5 mutagenesis: Expanding the toolbox for phage genome engineering. Frontiers in Microbiology 12:667332. doi: 10.3389/fmicb.2021.667332.
- Riggio, G. M., Q. Wang, K. E. Kniel, and K. E. Gibson. 2019. Microgreens-A review of food safety considerations along the farm to fork continuum. International Journal of Food Microbiology 290:76–85. doi: 10.1016/j.ijfoodmicro.2018.09.027.
- Rodríguez-Rubio, L., B. Martínez, A. Rodríguez, D. M. Donovan, and P. García. 2012. Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88 HydH5 virion-associated peptidoglycan hydrolase: Fusions, deletions, and synergy with LysH5. Applied and Environmental Microbiology 78 (7):2241–8. doi: 10.1128/AEM.07621-11.
- Romero-Calle, D., R. Guimarães Benevides, A. Góes-Neto, and C. Billington. 2019. Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics 8 (3):138. doi: 10.3390/antibiotics8030138.
- Ross, A., S. Ward, and P. Hyman. 2016. More is better: Selecting for broad host range bacteriophages. Frontiers in Microbiology 7:1352. doi: 10.3389/fmicb.2016.01352.
- Rustad, M., A. Eastlund, P. Jardine, and V. Noireaux. 2018. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synthetic Biology (Oxford, England) 3 (1):ysy002. doi: 10.1093/synbio/ysy002.
- Ryu, S. 2021. Grand challenges in phage biology. Frontiers in Microbiology 12:715039. doi: 10.3389/fmicb.2021.715039.
- Santos, S., E. Fernandes, C. M. Carvalho, S. Sillankorva, V. Krylov, E. Pleteneva, O. Shaburova, A. Nicolau, E. Ferreira, and J. Azeredo. 2010. Selection and characterization of a multivalent Salmonella phage and its production in a nonpathogenic Escherichia coli strain. Applied and Environmental Microbiology 76 (21):7338–42. doi: 10.1128/AEM.00922-10.
- Schmelcher, M., D. M. Donovan, and M. J. Loessner. 2012a. Bacteriophage endolysins as novel antimicrobials. Future Microbiology 7 (10):1147–71. doi: 10.2217/fmb.12.97.
- Schmelcher, M., F. Waldherr, and M. J. Loessner. 2012b. Listeria bacteriophage peptidoglycan hydrolases feature high thermoresistance and reveal increased activity after divalent metal cation substitution. Applied Microbiology and Biotechnology 93 (2):633–43. doi: 10.1007/s00253-011-3372-6.
- Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418 (6900):884–9. doi: 10.1038/nature01026.
- Shannon, R., D. R. Radford, and S. Balamurugan. 2020. Impacts of food matrix on bacteriophage and endolysin antimicrobial efficacy and performance. Critical Reviews in Food Science and Nutrition 60 (10):1631–40. doi: 10.1080/10408398.2019.1584874.
- Sharma, M., J. R. Patel, W. S. Conway, S. Ferguson, and A. Sulakvelidze. 2009. Effectiveness of bacteriophages in reducing Escherichia coli O157: H7 on fresh-cut cantaloupes and lettuce. Journal of Food Protection 72 (7):1481–5. doi: 10.4315/0362-028X-72.7.1481.
- Shen, Y., T. Köller, B. Kreikemeyer, and D. C. Nelson. 2013. Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. Journal of Antimicrobial Chemotherapy 68 (8):1818–24. doi: 10.1093/jac/dkt104.
- Shin, H., J.-H. Lee, H. Kim, Y. Choi, S. Heu, and S. Ryu. 2012. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PloS One 7 (8):e43392. doi: 10.1371/journal.pone.0043392.
- Shin, H., J.-H. Lee, H. Yoon, D.-H. Kang, and S. Ryu. 2014. Genomic investigation of lysogen formation and host lysis systems of the Salmonella temperate bacteriophage SPN9CC. Applied and Environmental Microbiology 80 (1):374–84. doi: 10.1128/AEM.02279-13.
- Shin, J., P. Jardine, and V. Noireaux. 2012. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synthetic Biology 1 (9):408–13. doi: 10.1021/sb300049p.
- Simmons, M., C. A. Morales, B. B. Oakley, and B. S. Seal. 2012. Recombinant expression of a putative amidase cloned from the genome of Listeria monocytogenes that lyses the bacterium and its monolayer in conjunction with a protease. Probiotics and Antimicrobial Proteins 4 (1):1–10. doi: 10.1007/s12602-011-9084-5.
- Soffer, N., J. Woolston, M. Li, C. Das, and A. Sulakvelidze. 2017. Bacteriophage preparation lytic for Shigella significantly reduces Shigella sonnei contamination in various foods. PloS One 12 (3):e0175256. doi: 10.1371/journal.pone.0175256.
- Son, B., M. Kong, Y. Lee, and S. Ryu. 2021. Development of a novel chimeric endolysin, Lys109 with enhanced lytic activity against Staphylococcus aureus. Frontiers in Microbiology 11:3490. doi: 10.3389/fmicb.2020.615887.
- Son, J.-S., S.-J. Lee, S. Y. Jun, S. J. Yoon, S. H. Kang, H. R. Paik, J. O. Kang, and Y.-J. Choi. 2010. Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Applied Microbiology and Biotechnology 86 (5):1439–49. doi: 10.1007/s00253-009-2386-9.
- Soni, K. A., and R. Nannapaneni. 2010. Removal of Listeria monocytogenes biofilms with bacteriophage P100. Journal of Food Protection 73 (8):1519–24. doi: 10.4315/0362-028x-73.8.1519.
- Sukumaran, A. T., R. Nannapaneni, A. Kiess, and C. S. Sharma. 2016. Reduction of Salmonella on chicken breast fillets stored under aerobic or modified atmosphere packaging by the application of lytic bacteriophage preparation SalmoFreshTM. Poultry Science 95 (3):668–75. doi: 10.3382/ps/pev332.
- Swift, S. M., B. S. Seal, J. K. Garrish, B. B. Oakley, K. Hiett, H.-Y. Yeh, R. Woolsey, K. M. Schegg, J. E. Line, and D. M. Donovan. 2015. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-Clostridium antimicrobial with improved thermostability. Viruses 7 (6):3019–34. doi: 10.3390/v7062758.
- Tao, P., X. Wu, W.-C. Tang, J. Zhu, and V. Rao. 2017. Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synthetic Biology 6 (10):1952–61.
- Timothy Kuan Ta Lu, M. S. K. B. S. Chevalier, J. W. Holder, G. J. McKenzie, and D. R. Brownell. 2011. Recombinant Phage and Methods. Inc., I. F. E. H. (Ed.).
- Torres‐Acosta, M. A., Clavijo, V.Vaglio, C. González, Barrios, A. F.Vives, Flórez, M. J. Rito, and Palomares, M. 2019. Economic evaluation of the development of a phage therapy product for the control of Salmonella in poultry. Biotechnology Progress 35 (5):e2852. ‐ ‐ ‐ doi: 10.1002/btpr.2852.
- Torres‐Acosta, M., A. González‐Mora, F. Ruiz‐Ruiz, M. Rito‐Palomares, and J. Benavides. 2020. Economic evaluation of M13 bacteriophage production at large‐Scale for therapeutic applications using aqueous Two‐Phase systems. Journal of Chemical Technology & Biotechnology 95 (11):2822–33. doi: 10.1002/jctb.6526.
- Vikram, A., J. Woolston, and A. Sulakvelidze. 2020. Phage biocontrol applications in food production and processing. Current Issues in Molecular Biology 40:267–302. doi: 10.21775/cimb.040.267.
- Vos, M. 2009. Why do bacteria engage in homologous recombination? Trends in Microbiology 17 (6):226–32. doi: 10.1016/j.tim.2009.03.001.
- WHO. 2016. Global Antimicrobial Resistance Surveillance System (GLASS): Guide to completing the GLASS implementation questionnaire. World Health Organization, https://apps.who.int/iris/handle/10665/251555
- WHO. 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. World Health Organization, https://apps.who.int/iris/handle/10665/311820
- WHO. 2020. Food safety. World Health Organization, https://www.who.int/news-room/fact-sheets/detail/foodsafety
- Wienhold, S.-M., J. Lienau, and M. Witzenrath. 2019. Towards inhaled phage therapy in Western Europe. Viruses 11 (3):295. doi: 10.3390/v11030295.
- Wittmann, J., C. Brancato, K. Berendzen, and B. Dreiseikelmann. 2016. Development of a tomato plant resistant to Clavibacter michiganensis using the endolysin gene of bacteriophage CMP1 as a transgene. Plant Pathology 65 (3):496–502. doi: 10.1111/ppa.12417.
- Woolston, J., A. R. Parks, T. Abuladze, B. Anderson, M. Li, C. Carter, L. F. Hanna, S. Heyse, D. Charbonneau, and A. Sulakvelidze. 2013. Bacteriophages lytic for Salmonella rapidly reduce Salmonella contamination on glass and stainless steel surfaces. Bacteriophage 3 (3):e25697. doi: 10.4161/bact.25697.
- Xu, Y. 2021. Phage and phage lysins: New era of bio-preservatives and food safety agents . Journal of Food Science 86 (8):3349–73. doi: 10.1111/1750-3841.15843.
- Yan, G., J. Liu, Q. Ma, R. Zhu, Z. Guo, C. Gao, S. Wang, L. Yu, J. Gu, D. Hu, et al. 2017. The N-terminal and central domain of colicin A enables phage lysin to lyse Escherichia coli extracellularly. Antonie Van Leeuwenhoek 110 (12):1627–35. doi: 10.1007/s10482-017-0912-9.
- Yang, H., S. B. Linden, J. Wang, J. Yu, D. C. Nelson, and H. Wei. 2015. A chimeolysin with extended-spectrum streptococcal host range found by an induced lysis-based rapid screening method. Scientific Reports 5 (1):17257. doi: 10.1038/srep17257.
- Yang, H., Y. Zhang, J. Yu, Y. Huang, X.-E. Zhang, and H. Wei. 2014. Novel chimeric lysin with high-level antimicrobial activity against methicillin-resistant Staphylococcus aureus in vitro and in vivo. Antimicrobial Agents and Chemotherapy 58 (1):536–42. doi: 10.1128/AAC.01793-13.
- Yeh, Y., F. De Moura, K. Van Den Broek, and A. De Mello. 2018. Effect of ultraviolet light, organic acids, and bacteriophage on Salmonella populations in ground beef. Meat Science 139:44–8. doi: 10.1016/j.meatsci.2018.01.007.
- Yehl, K., S. Lemire, A. C. Yang, H. Ando, M. Mimee, M. D. T. Torres, C. de la Fuente-Nunez, and T. K. Lu. 2019. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179 (2):459–469. e459. doi: 10.1016/j.cell.2019.09.015.
- Yeom, H., T. Ryu, A. C. Lee, J. Noh, H. Lee, Y. Choi, N. Kim, and S. Kwon. 2020. Cell-free bacteriophage genome synthesis using low-cost sequence-verified array-synthesized oligonucleotides. ACS Synthetic Biology 9 (6):1376–84. doi: 10.1021/acssynbio.0c00051.
- Yin, H., J. Li, H. Huang, Y. Wang, X. Qian, J. Ren, F. Xue, J. Dai, and F. Tang. 2021. Microencapsulated phages show prolonged stability in gastrointestinal environments and high therapeutic efficiency to treat Escherichia coli O157: H7 infection. Veterinary Research 52 (1):118. doi: 10.1186/s13567-021-00991-1.
- Yoichi, M., M. Abe, K. Miyanaga, H. Unno, and Y. Tanji. 2005. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. Journal of Biotechnology 115 (1):101–7. doi: 10.1016/j.jbiotec.2004.08.003.
- Yoong, P., R. Schuch, D. Nelson, and V. A. Fischetti. 2004. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. Journal of Bacteriology 186 (14):4808–12. doi: 10.1128/JB.186.14.4808-4812.2004.
- Żaczek, M., B. Weber‐Dąbrowska, and A. Górski. 2015. Phages in the global fruit and vegetable industry. Journal of Applied Microbiology 118 (3):537–56. doi: 10.1111/jam.12700.
- Zampara, A., M. C. H. Sørensen, D. Grimon, F. Antenucci, A. R. Vitt, V. Bortolaia, Y. Briers, and L. Brøndsted. 2020. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Scientific Reports 10 (1):12. doi: 10.1038/s41598-020-68983-3.
- Zhang, H., H. Bao, C. Billington, J. A. Hudson, and R. Wang. 2012. Isolation and lytic activity of the Listeria bacteriophage endolysin LysZ5 against Listeria monocytogenes in soya milk. Food Microbiology 31 (1):133–6. doi: 10.1016/j.fm.2012.01.005.
- Zhang, X., Y. D. Niu, Y. Nan, K. Stanford, R. Holley, T. McAllister, and C. Narváez-Bravo. 2019. SalmoFresh™ effectiveness in controlling Salmonella on romaine lettuce, mung bean sprouts and seeds. International Journal of Food Microbiology 305:108250. doi: 10.1016/j.ijfoodmicro.2019.108250.
- Zhou, Y., H. Zhang, H. Bao, X. Wang, and R. Wang. 2017. The lytic activity of recombinant phage lysin LysKΔamidase against staphylococcal strains associated with bovine and human infections in the Jiangsu province of China. Research in Veterinary Science 111:113–9. doi: 10.1016/j.rvsc.2017.02.011.
