Abstract
Thirty-six strains of lactobacilli isolated during the processing of traditional Lighvan cheese were analyzed biochemically and physiologically. The authors examined lactobacillus biochemical fingerprinting of selected isolates with a Phene-Plate-LactoBacilli (PhP-LB) system and determined their resistance to 15% salt and low temperature (4°C), their ability to produce acid, and their specific growth rate. Some strains showed qualities that might be promising for industrial-scale use. The results showed a high biodiversity among lactobacilli isolated from Lighvan cheese and offered a remarkable reservoir of ‘natural’ microbes. The potential of PhP-LB system for grouping and diffracting lactobacilli strains has been successfully demonstrated.
INTRODUCTION
Lighvan cheese is an Iranian variety manufactured from raw ewe or goat milk without the addition of starter cultures, according to the traditional method previously reported.Citation[1] In recent years, there has been a growing interest in genotypic and phenotypic studies on wild isolates from artisanal cheeses.Citation2–5 This information may also help prevent the extinction of a wide range of cheeses produced by different methods, whose typical features depend on many factors, such as local and regional traditions and also the indigenous microbial population present in raw milk, which is selected by the cheese-making environment.Citation[6] The extensive use of defined starters composed of a few selected strains has resulted in low strain variability.Citation[7] In order to expand the range of fermented milk products with different characteristics and enhanced market value, the biodiversity found in natural ecological niches should be exploited.Citation[8]
Non processed milk seems to be the main factor in producing the particular sensory properties of raw milk cheeses.Citation[9] Therefore, the addition of native cultures to pasteurized milk would not only permit the manufacture on an industrial scale of a uniform and safe product of constant quality, but also could help to protect the typical characteristics of traditional cheeses.Citation[10] In fact, wild LAB strains have been isolated and successfully applied in the manufacturing of dairy products to improve, for instance, the organoleptic properties and safety of the final product.Citation11–13 Lactobacilli are a heterogeneous group of lactic acid-producing anaerobic or microaerophilic bacteria.Citation[14] Given the great potential economic value of lactobacilli, isolation, counting, and identification are essential for quality assurance programs associated with the research, development, production and validation of the health or technological benefits of these bacteria.Citation[15]
The objective of this study was to conduct a preliminary physiological/biochemical characterization of the inter-species biodiversity of mesophilic lactobacillus species, which are dominant lactic acid bacteria (LAB) present during Lighvan cheese processing.Citation16 Citation17 Additionally, the capacity of the Phene-plate system to differentiate L. plantarum, L. paraplantarum, L. paracasei, and L. brevis strains was investigated. These strains had been previously isolated and identified by molecular methods (RAPD and 16S rRNA sequencing and species-specific PCR).Citation18
MATERIALS AND METHODS
Strains
Previously, 125 lactobacilli strains isolated from Lighvan cheeses were genotyped with the RAPD method, using two random sets of amplified primers, M13[1] and primer no. 4 of the Arsham RAPD kit.Citation1 Citation18 Thirty-six strains representative of each genotype were chosen for PhP fingerprinting and physiological/biochemical analysis.
Phenotyping: PhP Fingerprinting and Biochemical Characterization
After microscopic examination, Gram-positive and rod-shaped bacteria were subjected to a simple method for biochemical fingerprinting of lactobacilli, using the PhenePlate™ (PhP-LB) system, according to the manufacturer's instructions (PhPlate Microplate Techniques AB, Stockholm, Sweden). Briefly, the PhP system, modified for typing lactobacilli (PhP-LB), consists of 96-well microplates, each with four sets of 24 dehydrated reagents. In these reagents, 23 serve as discrimination tools for typing lactobacilli isolates from milk and cheese samples. The 24th well is a negative control. The lactobacilli to be tested were pre-cultivated on MRS agar (Merck, Darmstadt, Germany) plates. PhP suspending medium, with the indicator bromocresol purple, was dispensed into sterile capped vials: 5 mL per vial. The system is not very sensitive to bacterial concentration; thus, the amount of bacteria varied. However, the preferred number of colonies of a certain isolate (between 1 and 10) was transferred to a vial of suspending medium and left suspended for at least 30 min. The suspension was then poured into a sterile Petri dish, and 150 μL was dispensed by multichannel pipette in each well of rows A–B for the first isolate (C–D for the second, E–F for the third, and G–H for the fourth isolate). Plates were incubated at 37°C under anaerobic conditions for 3 days. Biochemical activity of the isolates (change of the indicator color) was read visually (revealed by a change to a darker color or black) and represented by a positive sign (+), while a negative sign (−) and a negative/positive sign (±) represented no change and weak change, respectively. Thus, the biochemical fingerprint for each isolate consisted of 24 numbers. Pairwise similarities between the biochemical fingerprints were calculated and expressed as a correlation coefficient, which yielded a similarity matrix. The similarity matrix was clustered according to the un-weighted pair group method by using unweighted pair group average linkage analysis (UPGMA) to yield a dendrogram.Citation14 Reproducibility was determined according manual instruction. Briefly, five bacterial strains were used as references and were tested by the system in order to establish intra-assay reproducibility.
Physiological Methods
The level of gas production from glucose was determined in MRS broth containing inverted Durham tubes; a catalase test was performed by using 3% H2O2 (hydrogen peroxide). Viability in challenge conditions was determined in 10 mL of MRS broth containing 5, 10, 15, 20, and 25% (w/vol) NaCl. Samples were inoculated (1% vol/vol) with overnight cultures and incubated for 48–72 h at 30°C. Conventional cultures in MRS broth were used as the control.Citation19 The specific growth rate of each isolate was determined in the logarithmic phase according to this formula:
Statistical Analysis of Phenotypic Traits
A matrix of data consisting of phenotypic traits from each microorganism was constructed. The data were examined with the MVSP Program version 2.0 (W.L. Kovach, Institute of Earth Studies, University College of Wales, Aberystwyth, UK). Clustering was achieved by UPGMA.Citation14
RESULTS AND DISCUSSION
Metabolic fingerprints of wild Lactobacilli strains selected from each genotype are shown in . All of the isolates—except strain M9, identified as L. brevis, and strains Y5 and Y13, identified as L. paraplantarum—were able to ferment lactose (lac+), which is undoubtedly a major technological trait in dairy manufacturing.Citation7 All members apart from M2 and R21, identified as L. brevis and L. paracasei, respectively, could ferment galactose. Sorbose, galacturonic-lacton, and rhamnose were not fermented by any strain (data not shown).
Table 1 Phenotypic characterization of lactobacilli isolates based on Phene-plate system
Many differences among strains regarding the capacity to ferment other sugars were found. Similar results have previously been reported,[7] in which there was much variation in the capacity to ferment carbohydrates among wild strains. Coeuret et al.Citation15 also demonstrated that the identification of lactobacilli isolates at the species level may be difficult because of considerable variation in fermentation profiles, especially for the so-called Lactobacillus plantarum group (L. plantarum, L. paraplantarum, and L. pentosus), the Lactobacilllus casei and Lactobacilllus paracasei groups (L. casei, L. rhamnosus, L. zeae, and L. paracasei), L. brevis, and L. buchneri.
Figure 1 The biochemical fingerprints of isolates. Clustering was performed by an UPGMA of Pearson product moment correlation coefficient. The vertical dotted lines indicate the similarity value of 92%.
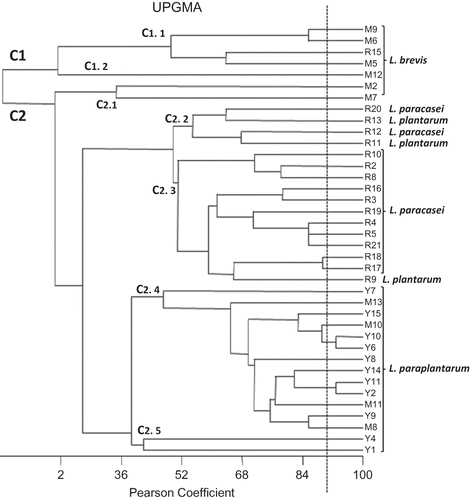
The biochemical fingerprints of all isolates were compared and UPGMA clustered, considering the reproducibility level of 0.92 for interassay analysis. Thirty-eight lactobacilli genotypes were distributed into 36 phenotypes showing a wide variation in sugar fermentation (Fig. 1). For example, none of L. paraplantarum strains showed the same fermentation pattern, and their abilities to ferment sugars, such as galactose, salicin, gloconate, amygdalin, manitol, and xylose, were diverse and strain-dependent.
Two major phenotypic groups (C1 and C2) and four subgroups, (C1.1 and C1.2) and (C2.1 and C2.2), were detected. L. brevis strains clustered into the C1 group and C2.1 subgroup and differed from other species in their ability to ferment arabinose and xylose and in their inability to ferment salicin. Other strains of L. paracasei, L. paraplantarum, and L. plantarum clustered into four subgroups of C2 from C2.2 to C2.5. All strains identified as L. paraplantarum grouped in the largest sub-cluster (C2.4) and also in the C2.5 sub-cluster differed from L. plantarum strains (clustered into C2.2 and C2.3) in their ability to ferment maltose and their inability to ferment palatinose, sucrose, melezitose, and tagatose. These sugars can be used as key sugars to differentiate those closely related species (L. plantarum and L. paraplantarum) that even 16S and 23S rRNA primers fail to discriminate.Citation22
The L. plantarum were also different from L. paracasei strains clustered into C2.2 and C2.3 in their ability to ferment sorbitol and their inability to ferment sucrose, trehalose, palatinose, and tagatose. As was shown in the dendrogram, L. brevis and L. paraplantarum were grouped into distinctive subgroups, but L. plantarum and L. paracasei showed a very similar sugar fermentation pattern and grouped into closer subgroups. Similar results have been obtained before by molecular techniques and genotypic characterization, in which these species also demonstrated a close genetic pattern.Citation1
The authors' results showed that PhenePlate™ technology offers an appropriately accurate distinction between different strains within a species, confirming previous resultsCitation14 that reported successful differentiating of lactobacilli strains isolates from Karst ewe's cheese with this technique. Our results showed an agreement between the phenotypic grouping patterns of isolates and RAPD clusters. This is the opposite of the results obtained by NigatuCitation23 and Botina et al.Citation24 who reported a disagreement between genetic and phenotypic patterns.
The physiological character of strains isolated during Lighvan cheese processing is shown in . Differences in acid-producing ability were found between genotypes, even within the same species; the decrease in pH from one strain to another ranged from 0.01 to 1.26 units. Some strains of L. paracasei (R3, R18, and R21) and L. brevis (M5 and M7) showed a higher acidifying ability than other strains.
Table 2 Physiological activities of lactobacilli isolates during Lighvan cheese maturing
Apart from R13 and M12 strains (identified as L. paracasei and L. brevis), all strains were able to coagulate milk and reduce its pH below 4 after 36 h incubation and with 10% v/v inoculation. Other researchers have also reported that Lactoacilli can metabolize lactose and produce acid similar to lactococci, albeit more slowly.Citation25 Citation26 All lactobacilli strains were able to grow at 4°C and in the presence of 5 and 10% NaCl (data not shown). Most of strains could grow in 15% NaCl but their growth was suspended in 20 and 25% NaCl (data not shown). The ability of lactobacilli to tolerate harsh conditions of low pH and high salt has been reported previously.Citation27 Citation28 The specific growth rate of lactobacilli was variable even among strains belonging to the same species. Strains M10, identified as L. paraplantarum, and R5 and R21, identified as L. paracasei, had the highest specific growth rates (approximately 0.7). In general, some strains in this study displayed promising properties, suggesting that they might be candidate strains to use as adjunct cultures in the manufacturing of cheeses or to use in the production of a new product. Among these, R4, R5, and R18 are the better candidates since they contribute efficiently to the removal of both lactose and galactose from milk, as evidenced by their higher specific growth rates; moreover, they demonstrate acceptable resistance in harsh conditions, and have suitable acid producing ability. Hence, products fermented with these strains would turn into more suitable food for the lactose-intolerant population and minimize the risks associated with galactose consumption.[7] Additionally, these strains showed resistance against low pH and could coagulate milk, reducing its pH by 3–4 units. This means they could be combined with high acid and nisin producers of lactococci. The application of L. casei together with Lactococcus strains with high acid production may positively affect cheese quality during the ripening period.Citation22 Although, some of L. brevis strains showed hopeful physiological properties, like the ability to ferment lactose and galactose, tolerate harsh conditions, and considerable acid production, they seemed to not be appropriate for use in culture.
These obligate heterofermentative strains may be associated with the generation of undesirable flavors, the formation of slits, and the decarboxilation of free amino acids to toxic amines.Citation29 Citation30 The role of lactobacilli in ripening of cheeses has not yet been satisfactorily resolved. However, some authors argue that selected strains could improve the flavor of cheeses made from pasteurized milk. The high degree of species and strain diversity found in the cheeses analyzed allows us to confirm that the addition of mixed native cultures and the starter to pasteurized milk, with an assumed influence on the organoleptic properties of the cheeses, would permit a uniform and safe product of consistent quality on an industrial scale. However, the careful choice of natural isolates is essential for the successful development of new starters.Citation10 Citation21
CONCLUSION
In this study, phenotypic properties of 38 lactobacilli genotypes, which had been identified in the authors' previous study, were analyzed by the Phene-plate system. This technique grouped them in 36 different phenotypes showing a wide phenotypic heterogeneity, as well as genotypic heterogeneity. Also, this method offered appropriate accurate distinction even among different lactobacillus strains within a species isolated during Lighvan cheese ripening. Analysis of metabolic activity and some technological traits of each phenotype, including growth rate and resistance levels, demonstrated that the industrial potential of some strain is very promising, especially R4, R5, and R18 strains since they showed higher growth rate, higher resistance to acid and NaCl, and ability of lactose and galactose consumption and ability of growth in low temperature. These strains might be used in adjunct starter culture but pilot-scale trials currently in progress are crucial. This study also highlights the importance of preserving lactobacilli diversity, which might be responsible of typical features of traditional Lighvan cheese.
REFERENCES
- Kafili , T. , Razavi , S.H. , Emam Djomeh , Z. , Salehi , G.R. , Álvarez-Martin , P. and Mayo , B. 2010 . Antibiotic resistance-susceptibility profiles of Lactobacillus strains from Lighvan, a traditional Iranian raw milk cheese . MilchSchaft , 65 : 559 – 562 .
- Erdogan , A. and Qurses , M. 2005 . Lactic acid bacteria isolating from blue mouldy Tulum cheese produced with Penicillium Roqueforti . International Journal of Food Properties , 8 : 405 – 411 .
- Scintu , M.F. and Piredda , G. 2007 . Typicity and biodiversity of goat and sheep milk products . Small Ruminant Research , 68 : 221 – 231 .
- Sengul , M. and Ertugay , M.F. 2006 . Microbiological and chemical properties of cheese helva produced in Turkey . International Journal of Food Properties , 9 : 185 – 193 .
- Nikolic , M. , Terzic-Vidojevic , A. , Jovcic , B. , Begovic , J. , Golic , N. and Topisirovic , L. 2008 . Characterization of lactic acid bacteria isolated from Bukuljac, a homemade goat's milk cheese . International Journal of Food Microbiology , 122 : 162 – 170 .
- Fortina , M.G. , Ricci , G. , Acquati , A. , Zeppa , G. , Gandini , A. and Manachini , P.L. 2003 . Genetic characterization of some lactic acid bacteria occurring in an artisanal protected denomination origin (PDO) Italian cheese, the toma piemontese . Food Microbiology , 20 : 397 – 404 .
- Sanchez , J.I. , Martinez , B. and Rodrıguez , A. 2005 . Rational selection of Leuconostoc strains for mixed starters based on the physiological biodiversity found in raw milk fermentations . International Journal of Food Microbiology , 105 : 377 – 387 .
- Tailliez , P. , Quenee , P. and Chopin , A. 1996 . Estimation de la diversite´parmi les souches de la collection CNRZ: Application de la RAPD a un groupe de lactobacilles . Lait , 76 : 147 – 158 .
- Grappin , R. and Beuvier , E. 1997 . Possible implications of milk pasteurization on the manufacture and sensory quality of ripened cheese: A review . Bulletin of the International Dairy Federation , 327 : 16 – 19 .
- Herreros , A. , Arenas , R. , Sandoval , M.H. , Castro , J.M. , Fresno , J.M. and Tornadijo , M.E. 2007 . Effect of addition of native cultures on characteristics of Armada cheese manufactured with pasteurised milk: A preliminary study . International Dairy Journal , 4 : 328 – 335 .
- Ayad , H.E. , Omran , N. and El Soda , M. 2006 . Characterisation of lactic acid bacteria isolated from artisanal Egyptian Ras cheese . Lait , 86 : 317 – 381 .
- Poveda , J.M. , Sousa , M.J. , Cabezas , L. and McSweeney , P.L.H. 2003 . Preliminary observations on proteolysis in Manchego cheese made with a defined-strain starter culture and adjunct starter (Lactobacillus plantarum) or a commercial starter . International Dairy Journal , 13 : 169 – 178 .
- Rilla , N. , Martinez , B. and Rodriguez , A. 2004 . Inhibition of a methicillin-resistant Staphylococcus aureus strain in Afuega'l Pitu cheese by the nisin Z-producing strain Lactococcus lactis subsp. lactis IPLA 729 . Journal of Food Protection , 67 : 928 – 933 .
- Majhenic , A.Ç , Lorberg , P.M. and Rogelg , I. 2007 . Characterization of the Lactobacillus community in traditional Karst ewe's cheese . International Journal of Dairy Technology , 60 : 182 – 190 .
- Coeuret , V. , Dubernet , S. , Bernardeau , M. , Gueguen , M. and Vernoux , J.P. 2003 . Isolation, characterization and identification of lactobacilli focusing mainly on cheeses and other dairy products . Lait , 83 : 269 – 306 .
- Abdi , R. , Shikh-Zeinoddin , M. and Soleimanian-Zad , S. 2006 . Identification of lactic acid bacteria isolated from traditional Iraninan Lighvan cheese . Pakistan Journal of Biology Science , 9 : 99 – 103 .
- Barouei , J. , Karbassi , A. , Goddusi , H.B. and Mortazavi , A. 2008 . Lactic microflora present in liqvan ewe's milk cheese . International Journal of Food Properties , 11 : 407 – 414 .
- Kafili , T. , Razavi , S.H. , Emam Djomeh , Z. , Salehi , G.R. , Álvarez-Martin , P. and Mayo , B. 2009 . Microbial characterization of Iranian traditional Lighvan cheese over manufacturing and ripening via culturing and PCR-DGGE analysis: Identification and typing of dominant lactobacilli . European Food Research and Technology , 229 : 83 – 92 .
- Kandler , O. and Weiss , N. 1986 . “ Genus Lactobacillus Beijerinck 1901, 212AL ” . In Bergey's Manual of Systematic Bacteriology , Edited by: Sneath , P.H.A. , Mair , N.S. , Sharpe , M.E. and Holt , J.G. 1209 – 1240 . Baltimore , MD : Williams & Wilkins .
- Ayad , E.H.E. , Nashat , S. , El-Sadek , N. , Metwaly , H. and El-Soda , M. 2004 . Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria . Food Microbiology , 21 : 715 – 725 .
- Heberta , E.M. , Raya , R.R. , Tailliezb , P. and De Gioria , C. 2000 . Characterization of natural isolates of Lactobacillus strains to be used as starter cultures in dairy fermentation . International Journal of Food Microbiology , 59 : 19 – 27 .
- Berthier , F. and Ehrlich , S.D. 1998 . Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region . FEMS Microbiology Letters , 161 : 97 – 106 .
- Nigatu , A. 2000 . Evaluation of numerical analyses of RAPD and API 50 CH patterns to differentiate Lactobacillus plantarum, Lact. fermentum, Lact. rhamnosus, Lact. sake, Lact. parabuchneri, Lact. gallinarum, Lact.casei, Weissella minor, and related taxa isolated from kocho and tef . Journal of Applied Microbiology , 89 : 969 – 978 .
- Botina , S.G. , Trenina , M.A. , Tsygankov , Y.D. and Sukhodolets , V. 2007 . Comparison of genotypic and biochemical characteristics of Streptococcus thermophilus strains isolated from sour milk products can cause sepsis and meningitis of newborns; S. mutanscan . Applied Biochemistry and Microbiology , 43 : 598 – 603 .
- Dagdemir , E. and Ozdemir , S. 2008 . Technological characterization of the natural lactic acid bacteria of artisanal Turkish white pickled cheese . Society of Dairy Technology , 61 : 133 – 140 .
- McDonald , L.C. , Fleming , H.P. and Hassan , H.M. 1990 . Acid tolerance of Leuconostoc mesenteroides and Lactobacillus plantarum . Applied and Environmental Microbiology , 56 : 2120 – 2124 .
- Hegazi , F.Z. 1984 . The microbial flora of salted raw milk . Systematic and Applied Microbiology , 5 : 527 – 533 .
- Haddadin , S.Y. 2005 . Kinetic studies and sensorial analysis of lactic acid bacteria isolated from white cheese made from sheep raw milk . Pakistan Journal of Nutrition , 4 : 78 – 84 .
- Ostliea , H.M. , Eliassenb , L. , Florvaag , A. and Skeie , S. 2004 . Phenotypic and PCR-based characterization of the microflora in Norvegia cheese during ripening . International Journal of Food Microbiology , 94 : 287 – 299 .
- McSweeney , P.L.H. , Fox , P.F. , Lucey , J.A. , Jordan , K.N. and Cogan , T.M. 1993 . Contribution of the indigenous microflora to the maturation of Cheddar cheese . International Dairy Journal , 3 : 613 – 634 .