ABSTRACT
Twenty-one strains of the genus Lactobacillus and the genus Pediococcus, isolated from Polish raw fermented meat products, were examined for the potential probiotic properties: resistance to simulated gastric and intestine conditions, safety assessment, and antimicrobial properties. Strains were resistant to gastric enzymes and low pH (3–6 log CFU/mL decrease) and intestinal enzymes and bile salts (1–3 log CFU/mL decrease). Most strains were resistant to gentamycin, streptomycin, vancomycin, tetracycline, ciprofloxacin, and kanamycin. Three of them (Lb. brevis BAL1, BAL10, and KL5) produced β-glucuronidase, which excludes them from qualifying as safe. Seven strains had the ability to produce bacteriocins or bacteriocin-like substances. Overall, strains Lb. brevis SCH6, Pd. pentosaceus BAL6, and KL14 revealed selected superior characteristics (resistance to the gastrointestinal conditions, safety assessment, and antimicrobial properties) as compared to the other LAB strains investigated, which made them a viable bioprotective culture that can be inoculated in raw fermented meat products as starter cultures.
Introduction
Fermented meat products are commonly considered safe for consumption, and the acidification by lactic acid starter bacteria is one of the main preserving factors. The most frequently isolated lactic acid bacteria (LAB) from dry sausages processed with different technologies are Lactobacillus sakei, Lb. curvatus, and Lb. plantarum.[Citation1,Citation2] The isolation and selection of LAB that can be used as starter cultures in meat fermentation present a considerable challenge to the standardization and management of quality of dry fermented sausage. The basic starter cultures used in the meat industry are selected strains of homofermentative lactobacilli and/or pediococci (LAB), and Gram-positive catalase–positive cocci (GCC), non-pathogenic, coagulase-negative staphylococci, and/or kocuriae.[Citation3]
Today, the modern meat industry has to ensure high quality, reduce variability, and enhance sensorial characteristics in raw fermented meat production. Regarding such situations, the use of selected starter cultures is important to produce the desired flavour and aroma compounds and extend the shelf life of the final product. The new idea is searching for indigenous LAB isolated from fermented meats, which are especially well adapted to the ecological conditions of specific meat fermentations, controlling the ripening processes, inhibiting the growth of spontaneous microorganisms, and possessing probiotic properties. As fermented meat products are processed without heating, they could be suitable products for assessing probiotic LAB as starter cultures.[Citation4]
According to the FAO/WHO Report,[Citation5] probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host”. They “should be safe and resistant to gastric juices and be able to grow in the presence of bile”. There is a series of in vitro tests such as acid and bile tolerance, antimicrobial production, etc. that, although requiring further refinement, are usually applied as a first approach for the selection of potentially probiotic microorganisms. Although LAB have a GRAS (Generally Recognized As Safe) status, they should be carefully examined for resistance to antibiotics and for the production of harmful metabolites.[Citation5]
LAB originating from fermented meats are specially adapted to the ecology of meat fermentation. The rapid production of lactic acid in those products is primarily responsible for the quality and safety of the product.[Citation1,Citation6] Nevertheless, outbreaks of serious sausage-borne gastrointestinal infections with pathogens such as verocytotoxic (shigatoxic) Escherichia coli (VTEC/STEC), Salmonella, and Listeria monocytogenes do occur regularly.[Citation7]
LAB produce an array of antimicrobial substances (such as organic acids, diacetyl, acetoin, hydrogen peroxide, reuterin, reutericyclin, antifungal peptides, and bacteriocins). Therefore, there is an increasing interest in LAB derived from meat that can be used as starter or adjunct cultures in dry sausage fermentation. Their ability to produce bacteriocins and non-proteinaceous low-molecular-mass antimicrobial compounds (mainly lactic acid and hydrogen peroxide) is of importance.[Citation8]
The aim of the study was to assess the antibiotic resistance, survival under simulated gastrointestinal conditions, and examine the antimicrobial properties of LAB strains isolated from Polish raw fermented meat products as candidates for probiotic strains that could be included in starter cultures for the development of novel meat products.
Material and methods
Bacterial strains – isolation, strain identification, and growth conditions
Nine samples of traditional organic raw fermented meat products (sausage, pork roast, gammon), obtained from three different parts of production, were collected. Meat products were produced by the Meat Processing Plant “Jasiołka” in Dukla, which raises meat from local organic farms. Ten grams of samples were transferred aseptically into 90 mL Ringer’s solution (BioMaxima, Poland) and homogenized thoroughly. Serial dilutions were made and spread plated on De Man, Rogosa, and Sharpe – MRS agar (Merck, Darmstadt, Germany). Twenty-one of LAB strains had previously been isolated and phenotypically identified as Lactobacillus or Pediococcus.[Citation9]
In this study, all 21 isolates were characterized biochemically and genotypically using 16S rDNA sequencing. Genomic DNA extraction was carried out using the reagent kit of Genomic AX Bacteria Mini Spin (A&A Biotechnology, Poland) according to the producer manual. The primer combination 27F/1492R (5’-AGA GTT TGA TCC TGG CTC AG-3’/5’- GGT TAC CTT GTT ACG ACT T-3’) was used for the amplification of 16S rDNA. 16S rDNA sequencing was performed with the superior strain screened from the study by the Illumina technique. The homology search was carried out by BLAST software. The GenBank accession numbers for reference 16S rDNA gene sequences and their similarity are represented in .
Table 1. Genetic identification of the tested lactic acid bacteria strains.
To perform antimicrobial assay, the pathogen/spoilage strains were used as indicators: Pseudomonas fluorescens, Proteus mirabilis, Escherichia coli, Bacillus subtilis, Enterococcus faecalis – laboratory isolates, Salmonella enteritidis ATCC 13076 and three Listeria monocytogenes strains – ATCC 7644, ATCC 19111, and ATCC 15313.
Commercial probiotic strain Lactobacillus plantarum 299v was used as a reference strain to compare the results.
All LAB strains and indicator bacteria were routinely propagated in MRS broth and Nutrient broth (Biokar Diagnostics, Noack, Poland), respectively. LAB strains were grown at 37°C for 24 h; L. monocytogenes strains were incubated for 48 h. The strains were maintained in a mixture of the same medium containing 20% of glycerol at 80°C until further use. Enumeration of LAB bacteria was performed using the spread-plate method onto MRS agar medium (BioMaxima, Poland) and incubated for 72 h at 30°C.[Citation10]
Studies on resistance to simulated gastric and intestine conditions
Survival of LAB strains was evaluated in a simulated gastrointestinal tract in vitro. The stress conditions were selected considering a previous work[Citation11] and suggestions of the Minekus et al. study.[Citation12] The simulated juices were prepared fresh daily. The composition of juices and the incubation time are shown in .
Table 2. Composition of juices and incubation time of the LAB strains tested in a simulated gastrointestinal tract.
Next, 1 mL of overnight cultures of LAB strains was removed and serially diluted in Peptone water (Noack, Poland) and spread-plated to determine the initial number. The rest of the cultures were centrifuged and washed twice with phosphate-buffered saline (PBS; BioMaxima, Poland). The pellets from bacteria cultures (concentration approx. 109–1010 CFU/mL) were treated with 5 mL of gastric juice and incubated for 2 h at 37°C. Decimal dilutions were made and 1 mL of aliquot was then spread-plated onto MRS agar. Then the left suspension was centrifuged, supernatant was removed, and then treated with 10 mL of intestinal juice and incubated for 3 h at 37°C. Decimal dilutions were made and 1 mL of aliquots was then spread-plated onto MRS agar. The experiment was performed in three replications.
Safety assessment
Antibiotic resistance
The tested strains were evaluated against gentamycin (0.016–256 µg/mL), streptomycin (0.016–256 µg/mL), ampicillin (0.016–256 µg/mL), vancomycin (0.016–256 µg/mL), tetracycline (0.016–256 µg/mL), ciprofloxacin (0.016–256 µg/mL), chloramphenicol (0.016–256 µg/mL), kanamycin (0.016–256 µg/mL), penicillin (0.002–32 µg/mL), azithromycin (0.0001–128 µg/mL), and erythromycin (0.016–256 µg/mL) using the E-test (HiMedia, India). One hundred microliter of the active bacterial suspension (0.5 McFarland) was spread evenly on the surface of the MRS agar plate. MRS has been chosen as the best medium for LAB growth. The antibiotics strips were placed on the plates and incubated anaerobically at 37°C for 48 h. The results were measured according to the instructions given by the manufacturer and compared with the minimum inhibitory concentration (MIC) values suggestions of Danielsen and Wind[Citation13] and EFSA.[Citation14]
Enzyme activities
Enzymatic profiles of the tested strains were determined using API ZYM (bioMérieux, France) under anaerobic incubation conditions in accordance with the manufacturer’s instructions.
Antimicrobial activities
The LAB strains were characterized for their antimicrobial properties relative to Gram-positive and Gram-negative indicator strains using the well diffusion method.[Citation15,Citation16] Antimicrobial activities of whole bacteria culture (WBC), cell-free supernatant (CFS), and neutralized, catalase-treated supernatant (NCS) were tested. Overnight WBC were vigorously mixed and used for the assay. The CFS was obtained as follows: overnight cultures were centrifuged (6000 × g, 10 min) and the supernatants were sterilized by filtration (0.2 µm). The NCS were obtained by adding the 1 M NaOH to the supernatants to obtain pH approx. 7.0 ± 0.2, treating with catalase (300 IU/ml, Sigma Aldrich) and filter sterilization (0.2 µm).
Petri dishes poured with Plate Count Agar (Merck) were inoculated with a 200 µl culture of indicator strain (concentration approx. 106 CFU/mL). Wells of 5.5 mm diameter were bored, and poured with WBC, CFS, and NCS in three replications. Plates were incubated at 37°C for 24 h and the diameters of the inhibition growth zones were measured. Antimicrobial activity was characterized and classified based on the inhibition growth zones diameters, and was described as slight (mm diameter), medium (4–8 mm), high (8–12 mm), and very high (>12 mm).
Amplification of reference genes
Antibiotic resistance genes for β-lactam (bla), erythromycin erm(A), tetracycline [tet(M), tet(S)], and gentamycin [aac(6′)-aph(2′′)] were amplified by PCR according to Nawaz et al.[Citation17] Primers used in this study are given in . The PCR reactions were provided as follows. The reaction mixtures (25 μl) contained 12.5 µl 2xPCR Master Mix (A&A Biotechnology, Poland), 0.1–1 µM of each primer and 10 pg – 1 µg bacterial DNA. DNA fragments were amplified in a thermal cycler up to 40 cycles by using annealing temperatures given in . PCR products were subjected to agarose gel electrophoresis in TAE buffer. Gels were visualized under UV transillumination.
Table 3. Nucleotide sequences of primer sets used for PCR amplification.[Citation25]
Statistical analysis
Statistical analysis was performed using STATISTICA15 (StatSoft Inc.). The data were analysed by analysis of variance (ANOVA).
Results and discussion
Isolation and identification of LAB bacteria
The studies identified 21 strains of LAB: 12 strains of the genus Lactobacillus and nine strains of the genus Pediococcus (). The following species have been assigned: Lb. plantarum (SCH1, SCH 2, SCH 3, SCH4, BAL7), Lb. brevis (SCH5, SCH6, BAL1, BAL10, KL3, KL5, KL7), and Pd. pentosaceus (BAL3, BAL5, BAL6, BAL9, KL6, KL9, KL11, KL13, KL14). All tested bacteria strains showed 97–100% similarity level in the nucleotide sequence of the 16S rDNA gene of the Lactobacillus or Pediococcus-type strain. The nucleotide BLAST analysis showed that all 21 strains belong to the phylum Firmicutes, class Bacilli, and family Lactobacillaceae.
According to Bromberg et al., Casaburi et al., and Ruiz-Moyano et al.[Citation18,Citation19,Citation20] the ripening process of fermented meat products is dominated by LAB, mainly represented by Lactobacillus genus and cocci. Also Vasilev et al.[Citation21] have found that LAB were dominant microbiota (more than 6 log CFU/g) in traditional Serbian dry fermented sausages. Other microorganisms, such as Micrococcus, Enterococcus, Pediococcus, and Leuconostoc, were found to be less relevant. The most frequent LAB species present in fermented meat processes are Lb. sakei, Lb. curvatus, and Lb. plantarum. However, the current study indicated that Lb. plantarum and Lb. brevis dominated fermented pork roast and gammon; in turn Lb. brevis and Pd. pentosaceus dominated fermented sausage and gammon.
Studies on resistance to gastrointestinal conditions
Probiotics have to overcome physical and chemical barriers to offer health benefits. It means that they have to tolerate gastric acid and bile in the gastrointestinal tract.[Citation22] About 2.5 L of gastric juice and 1.0 L of bile are secreted each day in the stomach.[Citation23] For most organisms gastric juice is harmful; however, some LAB can survive and grow at relatively low pH because they have a system that simultaneously transports lactic acid and protons to the cell’s exterior. Tolerance to gastric juice and bile salts became one of the most important selection criteria for probiotic microorganisms candidates.[Citation23]
The resistance of the tested bacteria strains to simulated gastrointestinal conditions is shown in . The initial number of tested bacteria cells was 9–10 log CFU/mL. After 2 h of the incubation in simulated gastric juice, a significant reduction (p < 0.05) in the number of bacterial cells by 3–6 log CFU/mL was observed. After another 3 h of the incubation in simulated intestinal juice, a significant reduction (p < 0.05) in the number of bacterial cells by 1–3 log CFU/mL occurred.
Table 4. Resistance of the tested bacteria strains to simulated gastrointestinal conditions.
As compared to the commercial probiotic Lb. plantarum 299v, the tested strains Lb. plantarum SCH1, SCH2, Lb. brevis SCH5, KL7, Pd. pentosaceus BAL6 survived significantly better (about 1 log CFU/mL higher) in the simulated gastrointestinal tract in vitro. De Vries et al.[Citation24] and Hamon et al.[Citation25] observed high survival rate of Lb. plantarum 299v at low pH (2.0). The authors[Citation25,Citation26] suggest that isolates of the Lb. plantarum species are characterized by innate resistance to gastric acids. The most sensitive strains to in vitro gastric and intestine conditions were Lb. brevis KL5 and BAL10. Jensen et al.[Citation23] found that strain Pediococcus pentosaceus Q3 tolerated gastric juice (pH 3.12) with no reduction of viability after 3-h incubation. In our study, it was confirmed that the higher the pH value of gastric juice, the less harmful is it to the LAB strains.
In the previous studies, Chen et al.[Citation27] evaluated LAB resistance to gastrointestinal conditions in vitro. They applied higher pH of gastric juice (pH = 3.0) than in this experiment (pH = 2.5) and showed higher survival rates. This indicates that the pH of the environment is a key factor in the survival of strains in the simulated gastrointestinal tract.
In addition, intestinal condition factors are crucial for bacteria survival. Agaliya and Jeevaratnam[Citation28] examined Lactobacillus plantarum strains, isolated from fermented idli batter, and found that the strains were able to resist the physiological bile salts concentration (0.3%). Conjugated bile salts are secreted into the small intestine for the absorption of dietary fat, vitamins, and other fat-soluble compounds[Citation29]. It has been reported[Citation30] that 0.3% concentration of the bile salts is considered to be critical for this kind of in vitro screening. In the current study, different resistance of the tested LAB strains to intestinal enzymes and bile salts contained in the intestinal juice has been shown. This resistance, with regard to some strains, could be due to bile salt hydrolase (BSH) activity that deconjugates bile salts.[Citation26] The hypothesis will be checked in a future study.
Safety assessment
Antibiotic resistance
Different susceptibility and resistance to antibiotics have been observed, depending on the bacteria strain (). The precise MIC values are shown in the online supplementary information (SI) (Supplement 1). In the phenotypic research, most strains showed susceptibility to ampicillin, chloramphenicol, penicillin, and erythromycin. According to Nawaz et al.[Citation17] Lactobacillus strains isolated from traditionally fermented products are susceptible to penicillin and erythromycin. LAB strains are reported to be resistant to chloramphenicol,[Citation31] but in our study none of the isolates were found resistant to chloramphenicol, which is consistent with the data reviewed by Sharma et al.[Citation32] In addition, the tested strains exhibited resistance to gentamycin, streptomycin, vancomycin, kanamycin, and ciprofloxacin. Resistance of Lactobacillus and Pediococcus species to vancomycin is considered as intrinsic.[Citation33] The vancomycin resistance occurring in the Lactobacillus genus is classified as intrinsic due to the construction of the peptidoglycan of bacterial cell walls.[Citation34] In another study,[Citation32] it has been observed that probiotic strains isolated from supplements and probiotic dairy products also possess resistance to vancomycin, kanamycin, and gentamycin.
Table 5. Susceptibility or resistance of the lactic acid bacteria strains to antibiotics.
The genetic analyses confirmed the presence of the gene coding resistance to β-lactams, tetracycline, erythromycin, and gentamycin. The vast amount of the tested strains revealed the presence of tet(M) gene, which is not necessarily connected with the phenotype study. In another study,[Citation17] it was observed the presence of tet(M) gene in Lb. plantarum, Lb. salivarius, Lb. animalis, and Lb. brevis strains and it was not dependent on the source of the isolates. According to EFSA,[Citation33] species of Lactobacillus plantarum are intrinsically resistant to streptomycin and this was also found in the current study. In a different report,[Citation14] it was claimed that the genes encoding for the resistance to tetracycline are not intrinsic in the Pediococcus pentosaceus species.
In the study of Agaliya and Jeevaratnam,[Citation28] the tested Lb. plantarum strains showed resistance to ciprofloxacin and gentamycin, and it was considered natural resistance to the above antibiotics. The food chain might be considered as one of the main routes of transmission of genes from human, animals, and bacteria, especially fermented products that are not heat treated before consumption, such as raw fermented meat products[Citation32]. However, translocation from the gastrointestinal tract into the systemic circulation has not been reported yet[Citation22]. Due to the prevalence of antibiotic resistance in the environment, gene transfer to other genera should be monitored.
Enzymatic profile
In all tested LAB strains, high activities of leucine arylamidase and valine arylamidase have been observed. The results of the enzymatic activity of the tested strains are presented in Supplement 2 (SI). None of the strains was found to produce α-chymotrypsin. The high activity of naphtol-AS-BI-phosphohydrolase was observed. No high activities of esterase, esterase lipase, and lipase were observed. LAB, particularly Lactobacillus and Lactococcus, are considered as weak lipolytic[Citation35]. Stoyanovski et al.[Citation36] found that strains isolated from Bulgarian meat product “Lukanka” had low and very low activity of esterase and lipase, respectively. In the current study, strains Lb. plantarum SCH1, SCH2, SCH3, SCH4, Lb. brevis SCH5, BAL1, BAL10, KL3, KL5, Pd. pentosaceus KL6, and KL9 had high activity of β-galactosidase, which is valuable in the food technology and in medicine.
β-glucuronidases liberate toxins and mutagens that have been glucuronated in the liver and excreted into the gut with the bile. This can lead to high local concentrations of carcinogenic compounds within the gut, thus increasing the risk of carcinogenesis. Also α-chymotrypsin, β-glucosidase, and N-acetyl-β-glucosaminidase activities may have negative effects in the colon[Citation37]. Among the tested strains, three of them (Lb. brevis BAL1, BAL10, and KL5) showed high activity of carcinogenic enzyme β-glucuronidase, which excludes them from qualifying as safe. None of the strains was found to produce α-chymotrypsin. The tested LAB strains isolated from raw fermented meat products were characterized by low enzyme activity as compared to the commercial probiotic strain Lactobacillus plantarum 299v.
Antagonistic activity
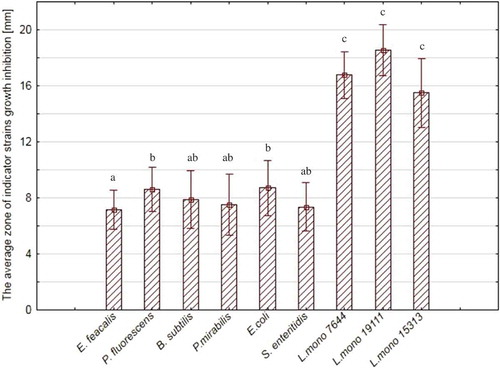
Antagonistic activity of the culture of the tested LAB strains was characterized by very high, high, and medium against the indicator strains, with the greatest zones of inhibition for L. monocytogenes ATCC 19111 (18.55 ± 1.8 mm) and the smallest for En. faecium (7.16 ± 1.1 mm). The results are presented in . It was found that the LAB strains were more active against Gram-positive indicator strains (p < 0.05). There was statistically significant differences (p < 0.05) between the antagonistic activity of WBC and NCS against Ps. fluorescens, B. subtilis, P. mirabilis, E.coli, L. monocytogenes ATCC 7644, L. monocytogenes ATCC 19111, and L. monocytogenes ATCC 15313.
Figure 1. Average antagonistic of WBC activity all of the lactic acid bacteria strains against indicator strains.
a, b, c – *statistically significant differences between the indicator strains (p < 0.05).
The CFS from the culture of the strains Pd. pentosaceus BAL9, Lb. brevis BAL10, and Pd. pentosaceus KL11 exhibited slight (1.9 ± 0.9 mm) antagonistic activity in vitro against E. coli, whereas the neutralized supernatant of strains Lb. brevis BAL10 and KL3 cultures exhibited antagonistic activity against S. enteritidis (3.5 ± 0.8 mm). In turn, neutralized supernatant from the culture of the strains Pd. pentosaceus BAL5, Lb. brevis SCH5, SCH6, Pd. pentosaceus KL13, and KL14 exhibited slight antagonistic activity against three tested L. monocytogenes strains, which suggests the ability of these LAB strains to produce bacteriocins or bacteriocin-like substances. Statistically significant differences between the antagonistic activity of bacteriocin-producing strains were also observed when S. enteritidis, L. monocytogenes ATCC 7644, and L. monocytogenes ATCC 15313 were tested. The results are presented in and .
Table 6. Antimicrobial activity of whole bacteria culture (WBC), cell free supernatant (CFS), and neutralized, catalase treated supernatant (NCS) against selected G(+) indicator strains.
Table 7. Antimicrobial activity of whole bacteria culture (WBC), cell free supernatant (CFS), and neutralized, catalase treated supernatant (NCS) against selected G(-) indicator strains.
The antagonistic activity of the LAB strains isolated from fermented meat products was comparable with that exhibited by probiotic microorganisms of human origin. Strong anti-listerial activity of LAB isolates from foods was observed by Sip et al., Cizeikiene et al., Choesri et al., and Wang et al.[Citation16,Citation38,Citation39,Citation40] Previous studies have shown that LAB strains can also produce bacteriocins that have different spectra of activity. Especially bacteriocins from class IIa have strong anti-listerial properties. Studies of other researchers confirm that the LAB strains exhibit low antagonistic activity against the strains of Enterococcus species.[Citation41] This indicator strain probably is immune for bacteriocins or bacteriocin-like substances produced by Lactobacillus strains isolated from meat products. Bacteriocin production is a common feature among Lactobacillus isolated from a variety of sources. It was observed that Lb. plantarum ULAG24, isolated from fermented cereal foods, inhibited Salmonella.[Citation42] In the Nghe and Nguyen study,[Citation43] bacteriocin produced by Pediococcus pentosaceus VTCC-B-601 effectively inhibited the growth of Salmonella typhimurium ATCC 19430 and Pseudomonas aeruginosa ATCC 27853.
Inhibition of indicator strains growth in most cases was attributed to the organic acids and other extracellular metabolites of LAB. Neutralization and catalase treatment of the supernatant reduced or eliminated the antagonistic activity against indicator strains in most cases. This indicates that the most important factor for inhibiting the growth of spoiler or pathogenic microorganisms is low pH of the environment. Moreover, natural antimicrobial agents are of great interest for the application in food preservation.[Citation43]
Conclusion
Most of the 21 studied LAB strains isolated from raw fermented meat products showed an adequate resistance to low pH and bile salt concentrations. Overall, strains Lb. brevis SCH6, Pd. pentosaceus BAL6, and KL14 reveal some superior characteristics (resistance to the gastrointestinal conditions, safety assessment, and antimicrobial properties) compared to the other LAB strains investigated. The summary of the potential probiotic characteristics of the tested LAB strains is shown in . The antimicrobial (especially anti-listerial) capacity plus the demonstrated probiotic properties made these strains a viable bioprotective culture that can be inoculated in raw fermented meat products as starter cultures. Further researches are required to study the in vivo probiotic properties (animal models and clinical tests) and technological usefulness.
Table 8. Summary of the probiotic characteristics of the tested LAB strains.
Supplement_2..docx
Download MS Word (15.1 KB)Supplement_1..docx
Download MS Word (15.9 KB)Acknowledgements
The authors would like to express their appreciation to Professor Zbigniew Dolatowski for support and constructive critiques during all study, and to Paweł Krajmas, who provided the material for study.
Supplemental data
Supplemental data for this article can be accessed on the publisher’s website.
References
- Bassi, D.; Puglisi, E.; Cocconcelli, P.S. Comparing Natural and Selected Starter Cultures in Meat and Cheese Fermentations. Current Opinion in Food Science 2015, 2, 118–122.
- Papamanoli, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E.; Kotzekidou, P. Characterization of Lactic Acid Bacteria Isolated from a Greek Dry-fermented Sausage in Respect of their Technological and Probiotic Properties. Meat Science 2003, 65, 859–867.
- Lücke, F.K. Utilization of Microbes to Process and Preserve Meat. Meat Science 2000, 56, 105–115.
- Kołożyn-Krajewska, D.; Dolatowski, Z.J. Probiotic Meat Products and Human Nutrition. Process Biochemistry 2012, 47, 1761–1772.
- FAO/WHO. Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation. 2001, 1–51.
- Stadnik, J.; Dolatowski, Z.J. Effect of Inoculation with Probiotics and Ageing Time on Selected Functional Properties and Oxidation of Proteins in Dry-cured Pork Loins. International Journal of Food Properties 2014, 17, 866–876.
- Moore, J.E. Gastrointestinal Outbreaks Associated with Fermented Meats. Meat Science 2004, 67, 565–568.
- Leroy, F.; Geyzen, A.; Janssens, M.; De Vuyst, L.; Scholliers, P. Meat Fermentation at the Crossroads of Innovation and Tradition: A Historical Outlook. Trends in Food Science & Technology 2013, 31, 130–137.
- Rzepkowska, A.; Zielińska, D.; Ołdak, A.; Kołożyn-Krajewska, D. Izolacja i identyfikacja szczepów bakterii fermentacji mlekowej pochodzących z wędlin surowo dojrzewających. In Bezpieczeństwo zdrowotne żywności. Aspekty mikrobiologiczne, chemiczne i ocena towaroznawcza; Stadnik J.; Jackowska I.; Eds.; Scientific Publishing PTTŻ: Kraków, 2015; 233–241.
- PN-ISO 15214:2002 Mikrobiologia żywności i pasz. Horyzontalna metoda oznaczania liczby mezofilnych bakterii fermentacji mlekowej. Metoda płytkowa w temperaturze 30°C.
- Rzepkowska, A.; Zielińska, D.; Kołożyn-Krajewska, D. Przeżywalność szczepów Lactobacillus wyizolowanych z żywności w warunkach modelowego przewodu pokarmowego. Żywność. Nauka. Technologia. Jakość 2015, 100, 42–52.
- Minekus, M.; Alminger, M.; Alvito, P.; Balance, S.; Bohn, T. et al. A Standardised Static in Vitro Digestion Method Suitable for Food: AnInternational Consensus. Food & Function 2014, 5, 1113–1124.
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus Spp. to Antimicrobial Agents. International Journal of Food Microbiology 2003, 82, 1–11.
- EFSA. Technical guidance. Update of the Criteria Used in the Assessment of Bacterial Resistance to Antibiotics of Human or Veterinary Importance. The EFSA Journal 2008, 732, 1–15.
- Hartmann, H.; Wilke, T.; Erdmann, R. Efficacy of Bacteriocin-containing Cell-free Culture Supernatants from Lactic Acid Bacteria to Control Listeria Monocytogenes in Food. International Journal of Food Microbiology 2011, 146, 192–199.
- Sip, A.; Więckowicz, M.; Olejnik-Shmidt, A.; Grajek, W. Anti-listeria Activity of Lactic Acid Bacteria Isolated from Golka, A Regional Cheese Produced in Poland. Food Control 2012, 26, 117–124.
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Cherie Millar, B.; Xu, J. Characterization and Transfer of Antibiotic Resistance in Lactic Acid Bacteria from Fermented Food Products. Current Microbiology 2011, 62, 1081–1089.
- Bromberg, R.; Moreno, I.; Zaganini, C.L.; Delboni, R.R.; de Oliveira, J. Isolation of Bacteriocin-Producing Lactic Acid Bacteria from Meat and Meat Products and its Spectrum of Inhibitory Activity. Brazilian Journal of Microbiology 2004, 35, 137–144.
- Casaburi, A.; Di Martino, V.; Ferranti, P.; Picariello, L.; Villani, F. Technological Properties and Bacteriocins Production by Lactobacillus Curvatus 54M16 and its Use as Starter Culture for Fermented Sausage Manufacture. Food Control 2016, 59, 31–45.
- Ruiz-Moyano, S.; Martin, A.; Benito, M.J.; Nevado, F.P. de Guia Cordoba, M. Screening of Lactic Acid Bacteria and Bifidobacteria for Potential Probiotic Use in Iberian dry Fermented Sausages. Meat Science 2008, 4, 199–208.
- Vasilev, D.; Aleksic, B.; Tarbuk, A.; Dimitrijevic, M.; Karabasil, N.; Cobanovic, N.; Vasiljevic, N. Identification of Lactic Acid Bacteria Isolated from Serbian Traditional Fermented Sausages Sremski and Lemeski Kulen. Procedia Food Science 2015, 5, 300–303.
- Vandenplas, Y.; Huys, G.; Daube, G. Probiotics: An Update. Jornal de Pediatria 2015, 91, 6–21.
- Jensen, H.; Grimmer, S.; Naterstad, K.; Axelsson, L. In Vitro Testing of Commercial and Potential Probiotic Lactic Acid Bacteria. International Journal of Food Microbiology 2012, 153, 216–222.
- De Vries, M.C.; Vaughan, E.E., Kleerebezem, M.; de Vos, W.M. Lactobacillus Plantarum: Survival,Functional and Potential Probiotic Properties in the Human Intestinal Tract. International Dairy Journal 2006, 16, 1018–1028.
- Hamon, E.; Horvatovich, P.; Marchioni, E.; Aoudé-Werner, D.; Ennahar, S. Investigation of Potential Markers of Acid Resistance in Lactobacillus Plantarum by Comparative Proteomics. Journal of Applied Microbiology 2014, 116(1), 134–144.
- Ferrando, V.; Quiberoni, A.; Reinheimer, J.; Suárez, V. Functional Properties of Lactobacillus Plantarum Strains: A Study in Vitro of Heat Stress Influence. Food Microbiology 2016, 54, 154–161.
- Chen, P.; Zhang, Q.; Dang, H.; Liu, X.; Tian, F.; Zhao, J.; Chen, Y.; Zhang, H.; Chen, W. Screening for Potential New Probiotic Based on Probiotic Properties and α-glucosidase Inhibitory Activity. Food Control 2014, 35, 65–72.
- Agaliya, P.J.; Jeevaratnam, K. Screening of Lactobacillus Plantarum Isolated from Fermented Idli Batter for Probiotic Properties. African Journal of Biotechnology 2012, 65, 12856–12864.
- Deshpande, K.G.; Dolas, C.B.; Chavan, N.S. Investigation of Tolerance of Lactobacillus Casei to the Presence of Acids, Bile Salts and Deconjugation of Bile Salts. International Journal of Current Microbiology and Applied Sciences 2014, 7, 600–612.
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-specific Probiotics Properties of Lactobacillus Fermentum, Lactobacillus Plantarum and Lactobacillus Brevis Isolates from Brazilian Food Products. Food Microbiology 2013, 36, 22–29.
- Zago, M.; Fornasari, M.E.; Carminati, D.; Burns, P.; Suarez, V.; Vinderola, G.; Reinheimer, J.; Giraffa, G. Characterization and Probiotic Potential of Lactobacillus Plantarum Strains Isolated from Cheeses. Food Microbiology 2011, 28, 1033–1040.
- Sharma, P.; Tomar, S.K.; Sangwan, V.; Goswami, P.; Singh, R. Antibiotic Resistance of Lactobacillus sp. Isolated from Commercial Probiotic Preparations. Journal of Food Safety 2016, 36, 38–51.
- EFSA. Scientific Opinion on the Safety and Efficacy of Pediococcus Pentosaceus (NCIMB 30044) as a Silage Additive for Animal Species. The EFSA Journal 2014, 12, 1–12.
- Gueimonde, M.; Sánchez, B.; de los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic Resistance in Probiotic Bacteria. Frontiers in Microbiology 2013, 4, 1–6.
- Zaręba, D. Profil kwasów tłuszczowych mleka sojowego fermentowanego różnymi szczepami bakterii fermentacji mlekowej. Żywność. Nauka. Technologia. Jakość 2009, 67, 59–71.
- Stoyanovski, S.; Gacoviski, Z.; Antonova-Nikolova, A.; Kirilow, N.; Ivanova, I.; Tenev, T.; Hadjinesheva, V. API ZYM Enzymatic Profile of Lactic Acid Bacteria Isolated from Traditional Bulgarian Meat Products “Lukanka”. Bulgarian Journal of Agricultural Science 2013, 19, 86–89.
- Heavey, P.M.; Rowland, I.R. Gastrointestinal Cancer. Best Practice & Research Clinical Gastroenterology 2004, 18 (2), 323–336.
- Cizeikiene, D.; Juodeikiene, G.; Paskevicius, A.; Bartkiene, E. Antimicrobial Activity of Lactic Acid Bacteria against Pathogenic and Spoilage Microorganism Isolated from Food and they Control in Wheat Bread. Food Control 2013, 31, 539–545.
- Choesri, D.; Rusmana, I.; Suwanto, A.; Mubarik, N.R. Characterization of Lactic Acid bacteria Isolated from Indonesian Fermented Fish (beksam) and their Antimicrobial Activity against Pathogenic Bacteria. Emirates Journal of Food and Agriculture 2013, 25, 489–494.
- Wang, G.; Zhao, Y.; Tian, F.; Jin, X.; Chen, H.; Liu, X.; Zhang, Q.; Zhao, J.; Chen, Y.; Zhang, H.; Chen, W. Screening of Adhesive Lactobacilli with Antagonistic Activity against Campylobacter Jejuni. Food Control 2014, 44, 49–55.
- Con, A.; Karasu, N. Determination of Antagonistic Starter Cultures for Pickle and Olive Fermentation Processes. Czech Journal of Food Science 2009, 3, 185–193.
- Oguntoyinbo, F.A.; Narbad, A. Multifunctional Properties of Lactobacillus Plantarum Strains Isolated from Fermented Cereal Foods. Journal of Functional Foods 2015, 17, 621–631002E
- Nghe, D.; Nguyen, T. Characterization of Antimicrobial Activities of Pediococcus Pentosaceus Vtcc-B-601. Journal of Applied Pharmaceutical Science 2014, 4, 61–64.
