ABSTRACT
The detection of milk fat adulteration with foreign fats/oils continues to be a challenge for the dairy industry as well as food testing laboratories. Recently, various chromatographic methods have been developed to detect a low level of vegetable oil in ghee but detection of animal body fat in ghee is still a challenge. In India, goat body fat (goat tallow) adulteration in ghee is a very serious problem. In the present investigation, a rapid species specific polymerase chain reaction (PCR)-based methodology has been standardised to detect goat tallow in heat clarified milk fat (ghee). This standardised PCR-based protocol did not show any false positive results in the genuine ghee (heat clarified butter fat) samples. Adulteration of ghee (both cow and buffalo) with goat tallow at the 10% level could be detected using the standardised PCR-based protocol. The protocol standardised is rapid and convenient to use.
Introduction
Food adulteration is a hidden evil of society, the main motive behind this crime is to earn more money. The problem of adulteration is not only affecting countries like India and Bangladesh, but also developed countries like UK and USA.[Citation1] Milk and dairy products are the easy and main targets of adulteration by unscrupulous traders. According to a report of FSSAI,[Citation2] 68% of the market milk samples of India were found to be below standards. In addition to milk, the high price milk products like ghee (clarified milk fat) are affected to the maximum extent. Ghee is one of the costliest edible fats which attract unscrupulous traders to add admixture of foreign fat to fetch more money. A wide range of adulterants were reported to be used in ghee like vegetable oil, mineral oil, animal body fat, paraffin etc. Among all these adulterants, body fat adulteration is one of the worst practices in Indian socio-economic culture from the religious and food habit points of views. Though vegetable oil is readily detected in ghee at very low levels,[Citation3–Citation5] detection of body fat adulteration in ghee is still a challenge[Citation6] as the analysis of physico-chemical characteristics alone is not very much helpful in detecting the presence of animal body fat in ghee. Similarly, some modern analytical techniques like derivative spectroscopy[Citation7] and the TLC-based method[Citation8] could not detect adulteration of ghee with animal body fat at a lower level. In addition to this, the said two techniques have only been used only in detecting the adulteration of cow ghee, whereas in India both cow milk and buffalo milk are commonly used for the preparation of ghee.
Goat tallow is easily available in India, as goat slaughter houses are readily available in every state of India. Previously, various methods like the opacity test,[Citation9] DTA technique,[Citation10] DSC, thin layer chromatography or GLC[Citation4]-based techniques etc., have been developed and used for the detection of goat body fat in ghee, but most of these techniques are time consuming and tedious. DNA is more resistant and thermo-stable than proteins and PCR amplifies small DNA fragments; also DNA could potentially be retrieved from any substrate because it is present in almost all cells of an organism.[Citation11] Therefore, DNA-based methodologies are gaining popularity for identifying the origin of food from animal sources (fish, milk, and meat). The DNA-based methodology for identification of species in milk and milk products,[Citation12] meat,[Citation13–Citation15] and fish[Citation16] has received important attention in the food industry due to high sensitivity and specificity. However, the DNA-based protocol to detect the addition of body fat and to ascertain the quality of clarified milk (ghee) fat has not been exploited till date.
Hence, the present study was planned to develop a PCR-based approach to detect the presence of goat tallow in ghee.
Materials and methods
Sample preparation and DNA isolation kit
Milk used for the preparation of respective ghee samples was collected from the Live-stock Research Centre of the National Dairy Research Institute, Karnal. Cow and buffalo ghee samples were prepared by the direct cream method.[Citation17] These samples were stored at refrigeration temperature till further analysis. Goat tallow was prepared from the adipose tissue collected from the slaughter house located at Karnal. Adipose tissues first washed thoroughly under running tap water and then heated over a direct flame at about 140°C till a transparent liquefied body fat was obtained. The liquid fat was filtered through muslin cloth followed by whatman filter paper No. 4 filter paper before its final use.
To prepare the adulterated ghee samples, pure ghee and goat tallow were heated to 60–70°C for 10 min then goat tallow was added to pure ghee @ 5%, 10% and 20%. The protocol of DNA isolation was essentially based on the method of Kumar et al. [Citation18] with small modifications. Forty five ml of molten fat, i.e. the ghee sample, was mixed with equal volume of ASL buffer followed by vortexing for 1 min and spinning for 5 min at 8000 rpm. The lower clear phase was transferred to a new tube and 3–4 ‘‘Inhibit EX Tablets’’ were added and contents were vortexed immediately for 1 min, followed by spinning for 5 min at 8000 rpm. The upper transparent solution (containing DNA) was transferred to a new 50 ml centrifuge tube followed by sequential addition of 1/10 volume of 3 M sodium acetate (5.3 pH) and 1 volume of ice cold absolute ethanol. The mixture was kept at −20° C for 45–60 min, spun for 2 min at 8000 rpm and thereafter the supernatant (SN) was discarded. The pellet or suspended material was re-suspended into 400 µl sterilised water and DNA was isolated using the protocol of the DNA isolation kit used in the study i.e. the QIAamp DNA stool mini kit. QIAamp DNA stool mini kit, ASL buffer and Inhibit EX tablets were purchased from (Quiagen,India, Pvt. Ltd.)
Isolation of DNA from blood
The genomic DNA isolated from the blood of cow, buffalo and goat by the method of Sambrook and Russel[Citation19] was procured from Core Lab of NBAGR, Karnal; for testing the cross amplification of selected goat specific primer (G) with cow or buffalo species
Goat specific primer (G)
A goat specific primer pair (G) with sequences Forward: TTCTTCAGGGCCATCTCATC and Reverse: GCGGATGCATGGTGAAAT resulting 294 bp amplicon for goat DNA, as reported by Kumar et al. [Citation20] was selected.
Polymer chain reaction conditions for the selected goat specific primer (G)
Polymerase Chain Reaction conditions were modified for the DNA isolated from ghee to make the reaction more specific and to avoid any cross reactivity with DNA isolated from cow and buffalo ghee. The reaction was carried out in 25 µl reaction volumes with about 25 ng genomic DNA using i-cycler (BioRAD, USA). The reaction mixture consisted of 200 µM each of dATP, dCTP, dGTP, dTTP, 3.5 mM MgCl2, 50 pmol of each forward and reverse primer (G), 1.0U Taq polymerase (Sigma-Aldrich®) and equal volume of corresponding Taq buffer. Amplification conditions were initial denaturation for 5 min at 94°C; followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min; and finally extension at 72°C for 5 min.
PCR product analysis
The PCR products were separated by electrophoresis on 2% agarose gels (with ethidium bromide staining) in parallel with a 100 bp DNA ladder (Fermentas, USA), at 90volts for 30 min.
Results and discussion
Selection and standardisation of goat specific primer
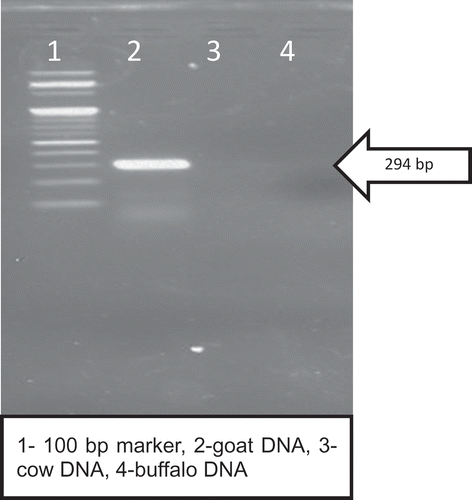
During initial phase of study, various goat specific primers selected from the literature, were used to amplify goat DNA isolated from goat blood. PCR conditions as described in the literature for each primer pair were used to amplify the DNA isolated from goat blood. To preclude the cross amplification, cow and buffalo DNA (from blood) was used for amplification. It was observed that most of the primers either failed to amplify goat DNA from goat blood or there was cross amplification with cow or buffalo DNA isolated from blood. Therefore, PCR conditions were standardised to make the primers highly goat specific, but most of the primers failed to become highly goat specific. Finally, the primer pair (G), which was used in this study, was able to amplify the DNA isolated from goat blood. It resulted in the desired and expected amplicon of 294bp from goat blood. The same primer G was checked for cross amplification with cow and buffalo blood DNA, wherein PCR conditions were kept same as reported by Kumar et al. [Citation20] It was observed that no PCR product corresponding to 294 bp amplicon was observed with genomic DNA isolated from blood of cow. However, with DNA isolated from buffalo blood, two bands were observed with size varying between 200 and 300 bp. The results of the study clearly demonstrated that PCR conditions described by Kumar et al. [Citation20] may have been optimised to work well in the case of meat samples used in their study. However, the same conditions could not work with DNA isolated from blood and showed cross reactivity of the primer with buffalo DNA. Therefore, the conditions of PCR were re-tuned to cater to prove the specificity of the selected set of primer
Standardisation of PCR conditions to increase the specificity of primer (G)
To make the primer G very specific to goat, different concentrations of MgCl2 were evaluated. It was observed that among various concentrations of MgCl2, only 3.5 mM concentration was found to be effective in providing the specificity of the primer in PCR reaction and thereby overcoming the problem of cross reactivity of primer G with buffalo blood DNA. It was also observed that beyond 3.5 mM concentration of MgCl2 no amplification was observed (). Our findings were in accordance with the results of Moreira et al.,[Citation21] who reported that too much MgCl2 is equally deleterious in most cases. In the case of excess Mg2+, the usual effect is a substantial increase in secondary products’ formation by non-specific priming. Hence, 3.5 mM concentration of MgCl2 was selected for setting up the PCR for primer G.
Table 1 Effect of different concentration of MgCl2 on the specificity of the goat specific primer G.
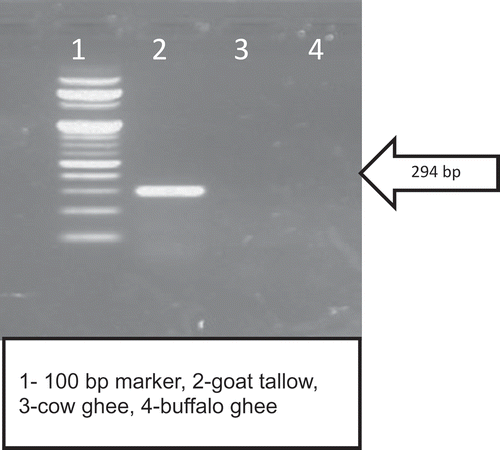
A fragment of 294 bp goat mitochondrial D-loop region was amplified for DNA isolated from goat blood (). It is evident that PCR product size of 294 bp was visible in 2% agarose gel but no amplification was recorded in case of cow or buffalo DNA. This clearly demonstrated that standardised PCR condition made primer G highly goat specific. Main challenge of this study was to isolate goat DNA from goat tallow. It is clear from that a 294 bp amplicon was visible in DNA isolated from goat tallow and there was no band visible in cow or buffalo ghee. Numerous factors have known to influence the outcome of successful PCR amplification for detection of adulteration in food e.g. quality of DNA, specificity of primers, etc.[Citation12] The main problem for developing the PCR-based approach to ascertain the purity of ghee are highly degraded DNA and recovery of DNA from fat matrices, as fat can potentially inhibit PCR amplification.[Citation22]The main challenge was to isolate amplifiable DNA for PCR reaction from fat matrices. In the initial phase of the study various DNA isolation kits for DNA isolation from heta clarified body fat and ghee or heat clarified milk fat. A protocol, earlier used by Noortiny et al. [Citation23] for DNA isolation from milk fat, were also tried but the said protocol was unable to isolate sufficient quantity of DNA from ghee for downstream application. The protocol used in this study was adopted from the protocol earlier used by Kumar et al. [Citation18] for isolation of DNA from olive oil, considering the fact that both oil and ghee are 99% triglycerides. However, the said protocol was not as such applicable for ghee so it was modified a little-bit so as to get sufficient quantity of DNA for downstream application. Hence, in the present work, 45 ml of ghee was used and during mixing of ASL buffer with ghee the temperature was maintained at 45°C. Maintenance of the temperature to 45°C enabled the fat to remain in liquid state and helped in obtaining the two different phases on high speed centrifugation. This treatment also allowed the ASL buffer to penetrate the fat matrix and liberated the degraded DNA in the lower phase. Hence, protocols used in the present study also yielded good quality of amplifiable DNA during PCR reaction as the absorbance ratio at 260/280 was 1.86 ± 0.12 for goat tallow. Earlier researchers[Citation24] reported that the column-based extraction protocol of DNA from food matrix was able to isolate sufficient quantity of DNA for downstream application, without the purification step of DNA. The same observations were reported by various researcher[Citation18,Citation25] for isolation of DNA from oil using the column-based commercial DNA isolation kit.
Detection of goat tallow in ghee by using goat specific PCR
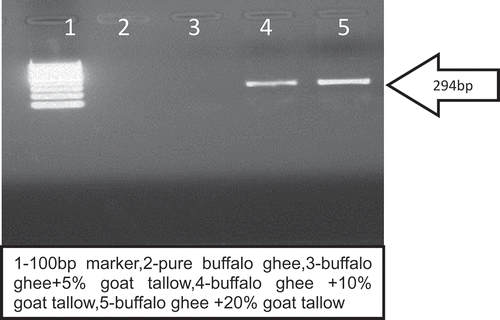
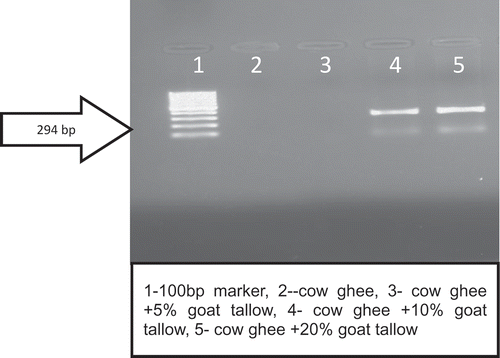
The oligonucleotide primer (G) was able to amplify the goat DNA even in the DNA isolated from ghee sample adulterated with goat tallow. It can be clearly observed that there was no amplification of DNA isolated from pure buffalo/cow ghee samples ( and ). It was also evident from the results of the study that in case of ghee (cow or buffalo) adulterated with goat tallow @ 5% no band was visible corresponding to a PCR product of 294 bp. Hence, this DNA-based method was able to detect 10% goat tallow addition in ghee.
Teletchea et al.[Citation11] reported that for species identification of highly processed food substrate small size DNA fragments were every time preferable for analysis. Hence, selected primer in this study yielded 294 bp amplicon, and was able to amplify degraded DNA from goat tallow (, and ). The findings of the present study are in the accordance with the findings of Hassan and Ali[Citation26] who were able to quantify goat DNA from milk powder using same set of primer. Hazra et al. [Citation27] reported that a simple species specific PCR-based approach is very successful for pin point the type of adulterants in milk or other dairy products. For checking the reproducibility of present assay we repeated the same protocol 10 times and observed the same result.
Conclusion
Present study was conducted to develop a simple PCR-based approach to detect goat tallow in ghee. DNA isolation from the fat-based food matrix is the pre-requisite for designing a PCR-based assay, thus the stool kit-based isolation technique, which is efficient in isolating ample quantity of DNA from ghee sample was used. The assay involves use of goat species specific primer, leading the detection of even up to 10% level of goat tallow adulteration in ghee ascertaining the type of body fat used to adulterate the milk fat.
Acknowledgement
The authors are very much thankful to the Directors of ICAR-NDRI, Karnal and ICAR-NBAGR, Karnal for providing facilities to carry out this research.
Funding
The authors are very much thankful to the Ministry of Food Processing-Government of India (MOFPI) for providing financial assistance.
Additional information
Funding
References
- Manning, L.; Soon, J.M. Developing Systems to Control Food Adulteration. Food Policy 2014, 49, 23–32.
- FSAAI. National Survey in India of Adulteration in Liquid Milk. http://www.fssai.gov.in/Portals/0/Pdf/sampleanalysed.2011(02-01-2012).pdf ( accessed Jan 12, 2016).
- Anupama, R.; Sharma, V.; Arora, S.; Lal, D.; Kumar, A.A. Rapid Reversed-Phase Thin Layer Chromatographic Protocol for Detection of Adulteration in Ghee (Clarified Milk Fat) with Vegetable Oils. Journal of Food Science and Technology 2013, 10, 2–6.
- Upadhay, N.; Detection of Vegetable Oil and Animal Body Fat in Ghee Using Solvent Fractionation Technique. PhD Thesis submitted to National Dairy Research Institute (Deemed University), Karnal, India. 2014.
- Anupama, R.; Sharma, V.; Arora, S.; Lal, D. Comparison of Rapid Reversed Phase High-Performance Liquid Chromatography (RP-HPLC) Method with Rapid Reversed Phase Thin Layer Chromatography Method for Detecting Vegetable Oils in Ghee (Clarified Milk Fat). International Journal of Food Properties 2016, 19, 1154–1162.
- Kumar, A.; Upadhyay, N.; Padghan, P.V.; Gandhi, K.; Lal, D.; Sharma, V. Detection of Vegetable Oil and Animal Depot Fat Adulteration in Anhydrous Milk Fat (Ghee) Using Fatty Acid Composition. MOJ Food Processing & Technology 2015, 1, 13–19.
- Jirankalgikar, N.M.; De, S. Detection of Tallow Adulteration in Cow Ghee Derivative Spectrophotometry. Journal of Natural Science Biology 2014, 5, 317–319.
- De, S.; Pankaj, N.; Jirankalgikar, N.M. Development of a Novel High Performance Thin Layer Chromatographicdensitometric Method for the Detection of Tallow Adulteration in Cow Ghee. JPC Journal of Planar Chromatography Modern TLC 2013, 26, 486–490.
- Singhal, O.P. Adulterants and Methods for Detection. India Dairyman 1980, 32, 771–774.
- Lambelet, P.; Singhal, O.P.; Ganguli, N.C. Detection of Goat Body Fat in Ghee by Differential Thermal Analysis. Journal of the American Oil Chemists Society 1980, 57, 364.
- Teletchea, F.; Maudet, C.; Catherine, H. Food and Forensic Molecular Identification: Update and Challenges. Trends in Biotechnology 2005, 7, 359–366.
- De, S.; Brahma, B.; Polley, S.; Mukherjee, A.; Banerjee, D.; Gohaina, M.; Singh, K.P.; Singh, R.; Datta, T.K.; Goswami, T.K. Simplex and Duplex PCR Assays for Species Specific Identification of Cattle and Buffalo Milk and Cheese. Food Control 2011, 22, 690–696.
- Doosti, A.; Dehkordi, P.G.; Rahimi, E. Molecular Assay to Fraud Identification of Meat Products. Journal of Food Science and Technology 2014, 51, 148–152.
- Şakalar, E.; Kaynak, A. Practical Molecular Detection Method of Beef and Pork in Meat and Meat Products by Intercalating Dye Based Duplex Real-Time Polimerase Chain Reaction. International Journal of Food Properties 2016, 19, 31–40.
- Ali, M.E.; Rashid, N.R.A.; Hamid, S.B.A.; Hossain, S.M.A.; Asing, A.; Hossain, M.A.M.; Zaidul, I.S.M. Development and Validation of Short-Amplicon Length PCR Assay for Macaques Meat Detection under Complex Matrices. International Journal of Food Properties 2017, 20, 231–245.
- Akasaki, T.; Yanagimoto, T.; Yamakami, K.; Tomonaga, H.; Sato, S. Species Identification and PCR-RFLP Analysis of Cytochrome B Gene in Cod Fish (Order Gadiformes) Products. Journal of Food Science 2006, 71, 190–195.
- De, S. Outlines of Dairy Technology; Oxford University Press: New Delhi, India, 2005; 441–443.
- Kumar, S.; Kahlon, T.; Chaudhary, S. A Rapid Screening for Adulterants in Olive Oil Using DNA Barcodes. Food Chemistry 2011, 127, 1335–1341.
- Sambrook, J.; Russell, D.W. Preparation and Analysis of Eukaryotic DNA. In Molecular Cloning: Alaboratory Manual; 3rd edn.; Cold spring Harbor Press: New York, USA, 1989; 631–632.
- Kumar, D.; Singha, S.P.; Singh, R.; Nagappa, S.K. A Highly Specific PCR Assay for Identification of Goat (Capra hircus) Meat. Small Ruminant Research 2011, 97, 76–78.
- Moreira, B.G.; You, Y.; Mark, A.B.; Richard, O. Effects of Fluorescent Dyes, Quenchers, and Dangling Ends on DNA Duplex Stability. Biochemical and Biophysical Research Communication 2005, 327, 473–484.
- Wilson, I.G.;. Inhibition and Facilitation of Nucleic Acid Amplification. Applied Environmental Microbiology 1997, 63, 3741–3751.
- Nooratiny, I.; Sahilah, A.M.; Alif Alfie, A.R.; MohdFarouk, M.Y. DNA Extraction from Ghee and Beef Species Identification Using Polymerase Chain Reaction (PCR) Assay. International Food Research Journal 2013, 20, 2959–2961.
- Maudet, C.; Taberlet, P. Detection of Cows’ Milk in Goats’ Cheeses Inferred from Mitochondrial DNA Polymorphism. Journal of Dairy Research 2001, 68, 229–235.
- Costa, J.; Isabel, M.; Joana, S.; Amaral, M.; Beatriz, P.; Oliveira, M.B.P.P. Detection of Genetically Modified Soybean DNA in Refined Vegetable Oils. European Food Research and Technology 2010, 230, 915–923.
- Hassan, K.I.; Ali, A.A. Evaluation of PCR Assay for Detection of Species Origin of Milk Powder. Journal of Biology & Life Sciences 2012, 201, 50–53.
- Hazra, T.; Sharma, V.; Sharma, R.; Arora, S. Simplex PCR Assay for Detection of Cow Milk Presence in Goat Milk. Indian Journal of Dairy Science 2016, 69, 621–625.