ABSTRACT
This study aimed to identify the dominant microbiota of crucian carp fillets during partial freezing (−3°C) and chilled (4°C) storage, and to correlate microbial changes with sensory scores and biogenic amines (BAs). The microbial communities of stored crucian carp fillets were identified by the high-throughput sequencing (HTS) technique of the V4 regions of the 16S ribosomal RNA gene. A total of 217,377 effective sequences and 690 different operational taxonomic units (OTUs) were identified and used in the analysis of microbial diversity. Culture-independent methods showed that the Pseudomonas were the predominant bacterial species of crucian carp fillets at the end of the storage. In addition, microbial enumeration also indicated relatively higher counts of Pseudomonas. Compared to chilled samples, partial freezing inhibited the growth of spoilage microorganisms during storage and extended the shelf-life of crucian carp fillets by 28 days based on the sensory analysis. However, there was no significant change in the composition of dominant species at the end of the shelf-life of crucian carp fillets.
Chemical compounds studied in this article: Tryptamine (PubChem CID: 150); Phenethylamine (PubChem CID: 1001); Putrescine (PubChem CID: 1045); Cadaverine (PubChem CID: 273); Histamine (PubChem CID: 774); Tyramine (PubChem CID: 5610); Spermidine (PubChem CID: 1102); Spermine (PubChem CID: 1103)
Introduction
The crucian carp (Carassius auratus) is one of the major freshwater fish species in China with an annual production of 2,767,900 tons in 2015.[Citation1] It has been widely consumed and cultured due to its desirable characteristics, such as high nutritional value, palatability, and reasonable domestic price. Fish is one of the most corruptible food products, and physical and chemical changes accompanied by microbial growth render the fish spoilage a very complex process. The undesirable microbial growth and metabolism are the primary causes of fish spoilage.[Citation2] Not all contaminated microorganisms are able to colonize the fish and grow to high numbers, only a particular part of these contaminants is responsible for the fish spoilage, known as “specific spoilage organisms” or SSOs.[Citation3]
Since the shelf-life of chilled crucian carp is relatively short, research on the application of new preservation methods is needed to extend the shelf-life of fresh fish. Partial freezing (superchilling) is a new kind of low-temperature preservation method intended to store a food product at 1–2°C below its freezing point, this can increase the shelf-life of foods by inhibiting most autolytic and microbial reactions. It has been reported that SSOs of fish are determined by raw materials used, processing parameters, packaging, and storage conditions of fish products.[Citation4] Recently, the spoilage bacteria and SSOs of chilled fish have been investigated by many researchers all over the word.[Citation5–Citation7] However, the SSOs of partial freezing fish and fish products have not been reported.
Culture-dependent methods can be used to investigate the SSOs of fish and fish products;[Citation8,Citation9] however, only less than 1% of the total bacterial population can be cultivated.[Citation10] Recently, high-throughput sequencing (HTS) technology, such as the 454 pyrosequencing of amplicons, has been widely used to analyse the microbial diversity of fish and fish products.[Citation11,Citation12]
In this study, a combination of HTS and microbial enumeration methods was used to investigate the microflora of crucian carp fillets stored at different temperatures. The objective of this study was to determine the dominant microbiota of the crucian carp fillets at the end of shelf-life. Additionally, we evaluated the effect of partial freezing on the composition of microbiota.
Materials and methods
Sampling and pretreatment
One hundred and eighty crucian carp (Carassius auratus) (370.0 ± 35.1 g body weight and 26.1 ± 1.5 cm body length) were purchased from an aquatic product wholesale market (Beijing, China) during the month of December 2015 and transported to the laboratory alive. The crucian carp were stunned, scaled, gutted, and filleted manually. Each fish was cut into two pieces, washed with running tap water, and then rinsed with cold sterile water. After that, the crucian carp pieces were packed in polyvinyl chloride bags and the fillets were divided into two groups and stored at 4°C and −3°C. The analysis of samples stored at 4°C and −3°C was conducted every 2 days and 5 days, respectively. However, the composition of bacterial communities was examined on 0, 6, and 12 days for chilled samples, and on 0, 35, and 40 days for partial freezing samples.
Sensory evaluation
The sensory evaluation of the fillets (fresh and cooked) was performed by nine trained panelists according to the method of Ojagh et al.[Citation13] with slight modification. The cooked crucian carp fillets were prepared by steaming for 5–10 min at 98°C. The colour, door, texture, and elasticity of raw fish muscle as well as flavour, door, and broth turbidity of cooked crucian carp fillets were evaluated by each evaluator on a 5-point scale, wherein 5 stands for the highest level and 1 the lowest. All the scores of separate aspects were summed to calculate the total sensory score. A total sensory score of 35 points represented absolutely fresh fish, and a score of 15 points considered as the limit for unacceptability of quality.
Determination of biogenic amines (BAs)
Extraction and analysis of BAs were performed by following the method of Hong et al.[Citation14] The identification and quantification of BAs were carried out using HPLC (LC-10AT series, Shimadzu, Tokyo, Japan) equipped with an SPD-10A (V) detector and a COSMOSIL 5C18-PAQ column (4.6 × 250 mm). Chromatographic separation was performed using a gradient elution program: 0 min, 50% B; 25 min, 90% B; 35 min, 90% B; and 45 min, 50% B. Ammonium acetate (0.1M) (solvent A) and acetonitrile (solvent B) were used as the mobile phases. In total, 50 µL of sample was injected at the flow rate of 0.8 mL/min and then detected by absorption at 254 nm.
Enumeration of microorganism
The crucian carp flesh (5 ± 0.05 g) was taken into stomacher bags with 45 mL of sterile saline (0.85%) and homogenized for 60 s using a stomacher (Masticator Basic L, S.A. Spain). Next, 100 μL of 10-fold serial diluted samples was spread on the surfaces of the culture medium. After 72 h of incubation at 30 ± 1°C, total viable counts (TVCs) were determined using plate count agar (PCA, Hai Bo Biological Technology Co. Ltd, Qingdao, China). Pseudomonas were counted on Pseudomonas Agar (CFC, Oxoid code CM0559, U.K.) with Pseudomonas CFC supplement (SR 0103E) at 20°C for 48 h. Aeromonas were determined on Aeromonas Medium Base (AMB, Oxoid code CM0833, U.K.) with supplement (SR 0136E), at 30°C for 48 h. Iron agar medium (IA, Hai Bo Biological Technology Co. Ltd, Qingdao, China) was used for the enumeration of H2S-producing bacteria and the plates were incubated at 20°C for 4 days.
DNA extraction
The total DNA extraction from the fish flesh was carried out according to the method of Xiao et al.[Citation15] with some modifications. The crucian carp flesh (6.5 g) was mixed with 20 mL of sterile saline (0.85%) and stirred for 15 min using an electric stirrer to let the bacteria suspend in physiological saline. The mixture was centrifuged at 200 g for 5 min. The supernatant was separated and centrifuged at 12000 g for 10 min. The pellet containing bacteria was collected and the total DNA was extracted from the pellet using a bacterial DNA extraction kit from Biomed Biological Technology Co., Ltd., (Beijing, China), according to the manufacturer’s instructions. The quality of DNA mixture was measured using 1.0% agarose gel electrophoresis.
Illuminahiseq2500 high-throughput sequencing
Primer pair, 515F (5ʹ-GTGCCAGCMGCCGCGGTAA-3ʹ) and 806R (5ʹ-GGACTACHVGGGTWTCTAAT-3ʹ) was used to amplify the V4 regions of bacterial 16S ribosomal RNA, giving PCR products of about 400–450 bp. PCR was carried out using a Phusion® High-Fidelity PCR Master Mix (New England Biolabs) with 6 μM of each primer, 5 ~ 10 ng of DNA template, and PCR-grade water. The PCR conditions were: pre-denaturation at 95°C for 1 min, then 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s, followed by a final extension step at 72°C for 5 min. The PCR products were stained with 1×loading buffer (contained SYB green), detected on a 2% agarose gel under UV light, and then purified by the Qiagen Gel Extraction Kit (Qiagen, Germany). Construction of sequencing libraries was performed using Seq® DNA PCR-Free Sample Preparation Kit (Illumina, USA). The quality of the library was verified using the Qubit@ 2.0 Fluorometer (Thermo Scientific) and the Agilent Bioanalyzer 2100 system. The bands that passed the quality check were sequenced on an IlluminaHiSeq2500 PE250 platform by Novogene Company (Beijing, China).
Sequences data preparation and analysis
After sequencing on the IlluminaHiSeq2500 PE250 platform, all sequences were sorted according to the unique barcode tagged to each sample. The PCR primer and barcode were trimmed from the sequences and resulted in paired-end (PE) reads. Raw tags were obtained by combination of PE reads using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/) and then sequences were quality filtered by QIIME 1.7.0 software (http:qiime.org/scripts/split_libraries_fastq.html). Raw tags were compared with the reference database (Gold database, http://drive5.com/uchime/uchime_download.html) and then chimera sequences were removed using the UCHIME algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html) to obtain effective tags. The effective sequences were then clustered into operational taxonomic units (OTUs) at 97% sequence similarity using Uparse v7.0.1001 (http://drive5.com/uparse/). According to the algorithm rule, the representative sequences of OTUs are the sequences of the highest frequency. The RDP Classifier v2.2 (http://sourceforge.net/projects/rdp-classifier/) and the GreenGene database (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) were used to classify the representative sequences of OTUs phylogenetically, with a confidence of 0.8. To determine the richness, diversity, and sequencing depth of bacterial communities, Chao1 value and ACE, Shannon and Simpson indices, and Good’s coverage were calculated using QIIME (v1.7.0) software. All these indices are related to Alpha diversity.
Statistical analysis
All the analyses of crucian carp fillets were repeated in triplicates, except for bacterial enumeration analyses, which were carried out in duplicate. Statistical difference between means was determined using the least significant difference (LSD) method and the significance at p < 0.05 was evaluated. Statistical analyses were performed using the SAS software (SAS Institute Inc., 2008).
Results and discussion
Sensory analysis
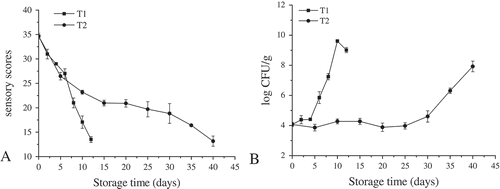
The changes in sensory scores of crucian carp fillets during chilled and partial freezing storage are shown in . The sensory scores of all the samples decreased significantly (p < 0.05) as the storage time increased, indicating the quality deterioration. However, a higher score was obtained for partial freezing storage samples than the chilled samples at the same storage period. Taking a total sensory score of 15.0 as the limit for unacceptability for quality, the fillets stored at 4°C and −3°C were inedible after 12 and 40 days of storage, respectively. Similar results were reported by Parlapani and Boziaris[Citation16] and it could be concluded that partial freezing could reduce the sensory changes in crucian carp fillets effectively.
Biogenic amines
The changes of eight BAs in crucian carp fillets stored at chilled and partial freezing temperatures are shown in . The initial values of tryptamine (TRM), phenylethylamine (2-PHE), putrescine (PUT), cadaverine (CAD), histamine (HIM), tyramine (TYM), spermidine (SPD), and spermine (SPM) were 3.12, 3.95, 5.12, 0.19, 6.02, 20.4, 5.87, and 4.68 mg/kg, respectively. The concentration of BAs of crucian carp fillets stored at −3°C was lower than those of chilled storage samples at the same storage period. Ozogul et al.[Citation17] stated that storage temperature is the most important factor affecting the production of BAs. TYM was the most abundant amine in all the samples. The concentration of TYM varied from 13.74 to 41.86 mg/kg in fillets of 4°C, and 22.10 to 31.21 mg/kg in samples stored at −3°C. SPD and SPM are natural polyamines in fish muscle[Citation14] and they fluctuated in two storage conditions in the present study. The PUT and CAD levels of chilled samples remained lower for 10 days, but increased significantly at day 12, whereas partial freezing fillets showed negligible fluctuations. Previous studies have reported that PUT and CAD are spoilage indicators[Citation18] of common carp stored at 20 and 0°C[Citation19] and carp flesh.[Citation20] In the current study, PUT and CAD are not suitable indices for the quality assessment of crucian carp fillets stored at −3°C. The HIM value of samples stored at 4°C was relatively higher than partial freezing samples and it gradually increased with time, although a slight fluctuation was observed during the first 10 days. In this study, the concentration of HIM is quite lower than that of crucian carp reported by Li et al.[Citation21]This might be due to the spoilage bacteria influenced by geographical origin and other unknown factors.
Table 1. Changes in biogenic amines content of crucian carp fillets stored at 4°C (CS) and −3°C (PFS).
Enumeration of microorganisms
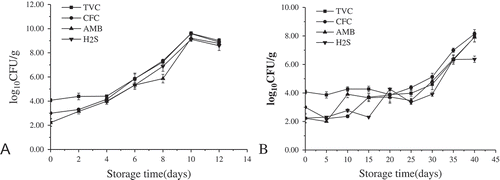
The changes in TVC of crucian carp fillets during chilled and partial freezing storage are shown in . Partial freezing samples had lower TVC counts (p < 0.05) compared with samples stored at 4°C in the same storage period. The origin TVC (4.07 log cfu/g) obtained in this study is comparable with the initial microbial load of rainbow trout fillets reported by Chytiri et al.[Citation21] The chilled storage samples showed a gradual increase of TVC values. However, the TVC values of the samples stored at −3°C increased slowly during the first 30 days and a sudden increase was observed after 30 days. This indicated that partial freezing had effective inhibitory effects on growth of the microbiota, but the destruction of tissue by tiny ice crystals might have accelerated the growth of the bacteria at the end of the shelf-life. The maximum acceptable level of TVC is 7.0 log cfu/g for freshwater and marine fish.[Citation23] The crucian carp fillets of chilled storage reached 7.27 log cfu/g after 8 days and the fillets of partial freezing storage reached 6.31 log cfu/g during 35 days. The sensory evaluation indicated that the products were not suitable for consumption on the 12th and 40th days for crucian carp fillets stored at 4°C and −3°C, in which the TVC levels were 9.01 and 7.92 log cfu/g, respectively. Obviously, the shelf-life of the samples depended on the microbial analysis.
The initial TVC was 4.07 log cfu/g, while Pseudomonas, Aeromonas, and H2S-producing bacteria population was 3.00, 2.23, and 2.24 log cfu/g (), respectively. The population of Pseudomonas was higher than Aeromonas and H2S-producing bacteria in chilled samples, and followed the same trend as TVC during the entire storage. However, in partial freezing samples, a fluctuation was observed for each microorganism before the 25th day, after which they followed the same trend as TVC and the population difference was not significant.
Relationship of BAs and microbial flora
The regression correlation coefficients for BAs and microbial flora of crucian carp fillets stored at 4°C and −3°C are illustrated in . Almost no significant correlations were shown between the TRM, 2-PHE, CAD, HIM, TYM and TVC, Pseudomonas, H2S-producing bacteria and Aeromonas in all samples, except for PUT and SPM in chilled samples and SPD in partial freezing samples. SPD and SPM exhibit significant correlations with microbial flora; however, the reason for its change is not completely clear.[Citation24] In addition, TVC and Aeromonas showed significant correlation(p < 0.05)with PUT in chilled samples. It is reported that Pseudomonas and Aeromonas are responsible for the accumulation of HIM and other related BAs such as PUT and CAD;[Citation25] however, HIM only increased after samples were considered spoilage for a long time.[Citation26,Citation27]
Table 2. Regression correlation coefficients between biogenic amines and microbial flora of crucian carp fillets stored at 4°C (CS) and −3°C (PFS).
Sequencing analysis
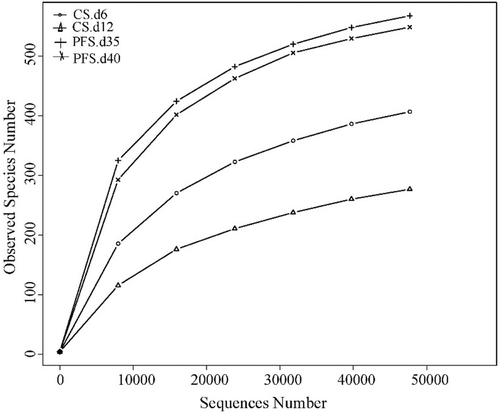
A total of 217,377 effective sequences (109,034 from chilled storage samples and 108,343 from partial freezing storage samples) with an average sequence length of 253 bp were recovered using the IlluminaHiSeq2500 platform. However, the composition of bacterial communities on day 0 was not detected through high-throughput sequencing, indicating the lower bacterial population in fresh crucian carp. The OTUs were clustered at 97% effective sequences similarity. As shown in , OTUs of the samples stored at 4°C and −3°C were 684 and 1117, respectively. The results demonstrated that partial freezing samples had higher microbial richness and diversity. The Good’s coverage returned values were above 99% in all samples, indicating a good sampling completeness. Compared to the observed number of OTUs, richness estimators (Chao1 and ACE) had higher values, suggesting additional bacterial phylotypes in the samples. Partial freezing samples were characterized as high in richness than chilled samples, as indicated by the high Shannon and Simpson values of partial freezing samples. The rarefaction curves of alpha diversity, based on the OTUs (), showed that all the samples tended to approach the saturation plateau at 4000 sequences. This indicated that the data size of the sequences tended to be more rational and can cover almost all the microbial communities.
Table 3. Comparison of phylotype coverage and alpha diversity estimation of the 16S rRNA gene libraries by sequencing on an IlluminaHiSeq2500 platform in crucian carp fillets stored at 4°C (CS) and −3°C (PFS).
Bacterial communities in crucian carp fillets
The composition and relative abundance of bacterial communities of crucian carp fillets were quantified using the HTS method. As shown in , all the samples were dominated by four phyla at different times of storage. Of the top 10 phyla, Proteobacteria followed by Firmicutes dominated the microbiota of crucian carp fillets throughout the storage. Proteobacteria occupied the overwhelming majority (with a relative abundance ranging from 77.6 to 93.4 %) in the samples under different storage conditions. Within Proteobacteria, the dominant phylum was Firmicutes and accounted for 5.42–13.18% in chilled and partial freezing samples. However, the relative abundance of Firmicutes in partial freezing samples was higher than the chilled fillets, which was negatively correlated with Proteobacteria. However, other phyla, such as Bacteroidetes, Acidobacteria, and Actinobacteria, were present at low percentages.
Figure 4. Relative abundance of microbiota based on phylum level (A), family level (B), and genus level (C) in crucian carp fillets stored at 4°C (CS) and −3°C (PFS).
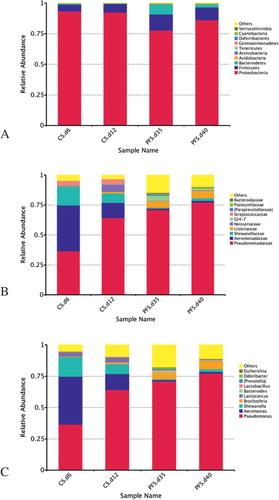
The top 10 families of chilled and partial freezing samples are shown in . Pseudomonadaceae and Aeromonadaceae were the predominant bacterial families (accounting for 36.61 and 38.19%, respectively) in the chilled samples on day 6, whereas Shewanellaceae accounted for 15.51%. Pseudomonadaceae (70.92%) followed by Listeriaceae (5.43%) was the prevalent population in partial freezing samples on day 35. A large shift was observed in the microbial communities of chilled samples on day 12, in which Pseudomonadaceae reached 64.19% and became the predominant bacterial family while the proportion of Aeromonadaceae and Shewanellaceae decreased to 12.68 and 7.88%, respectively. This may be attributed to the inability of the microbiota to compete with Pseudomonadaceae, which has the ability to grow well under anaerobic conditions,[Citation28] and the selective action of storage conditions. In comparison, the composition of bacterial communities on day 40 was similar to that on day 35. Pseudomonadaceae (77.27%) was significantly (p < 0.05) higher than any other microbiota at the end of the crucian carps’ shelf-life. In addition to Pseudomonadaceae, Listeriaceae was also the dominant family and accounted for 6.24% of the bacterial communities.
To analyse and compare the bacterial composition and community dynamics of crucian carp fillets under different storage conditions, a hierarchical clustering heat-map () was constructed at the genera level based on the relative abundance of the top 35 abundant phylotypes of all the samples. The results showed that the samples segregated into two clusters: one cluster was composed of chilled samples while the other cluster was formed by the two samples stored at −3°C. It was observed that the greater the relative abundances in the genera, the deeper the colour in each row of the heat-map. shows the abundances of the top 10 bacterial species at the genus level in four samples.
Figure 5. Relative abundance of microbiota at the genus level. Rows in the heat-map represent different genus-level phylotypes, while columns represent different crucian carp samples. The color intensity is proportional to the abundance of OTUs in each row (CS: chilled storage; PFS: partial freezing storage).
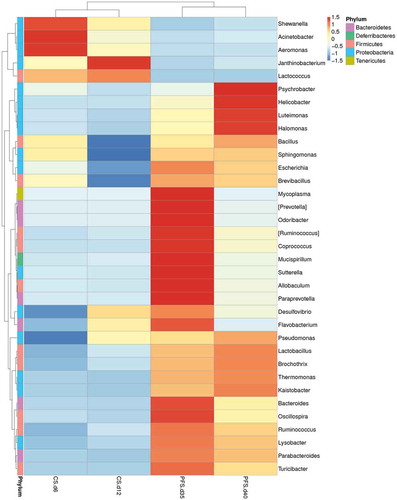
As shown in , Aeromonas (38.18%) and Pseudomonas (33.61%) were the most dominant genera detected on day 6, whereas Shewanella accounted for 15.55%. Compared to chilled samples on day 6, a higher abundance of Pseudomonas genera was observed in chilled samples on day 12 and all the partial freezing samples. The result is in accordance with the heat-map, in which a deeper colour of Pseudomonas was noticed in rows of those samples. Aeromonas and Shewanella also showed higher relative abundance (15.55%) and obtained the deepest colour in the row of chilled samples of day 6 than any other samples.
At the end of the storage period, Pseudomonas dominated the spoilage, which is in accordance with the previous scientific literature.[Citation4,Citation29–Citation31] These organisms may contribute much to the spoilage of chilled and partial freezing fillets. However, the second most common microbiota in partial freezing samples identified by microbial enumeration and the culture-independent method was Aeromonas and Shewenella, respectively, indicating the boundedness of the selective medium.
Conclusion
In this study, HTS showed that Pseudomonas followed by Aeromonas was the most abundant genera of chilled crucian carp fillets. Pseudomonas and Brochothrix were the dominant genera of partial freezing samples. However, Pseudomonas occupied the overwhelming majority in all the samples at the end of crucian carp fillets’ shelf-life and it was in accordance with the result of the culture-dependent approach. Partial freezing had no effect on the predominant microbiota. However, it inhibited the growth of the microbiota and extended the shelf-life of partial freezing crucian carp fillets. Further studies are needed to investigate the spoilage ability of Pseudomonas and determine the SSOs of chilled and partial freezing crucian carp fillets.
Funding
This study was supported by the earmarked fund for China Agriculture Research System (CARS-46), National Natural Science Foundation of China (award no.31471683), and Beijing Natural Science Foundation (award no. 6152017).
Additional information
Funding
References
- Bureau of Fisheries of the Ministry of Agriculture. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2016; 31 pp.
- Gram, L.; Huss, H.H. Microbiological Spoilage of Fish and Fish Products. International Journal of Food Microbiology 1996, 33(1), 121–137.
- Dalgaard, P. Qualitative and Quantitative Characterization of Spoilage Bacteria from Packed Fish. International Journal of Food Microbiology 1995, 26(3), 319–333.
- Gram, L.; Dalgaard, P. Fish Spoilage Bacteria: Problems and Solutions. Current Opinion in Biotechnology 2002, 13(3), 262–266.
- Parlapani, F.F.; Meziti, A.; Kormas, K.A.; Boziaris, I.S. Indigenous and Spoilage Microbiota of Farmed Sea Bream Stored in Ice Identified by Phenotypic and 16S Rrna Gene Analysis. Food Microbiology 2013, 33(1), 85–89.
- Zhang, Y.M.; Li, Q.; Li, D.P.; Liu, X.C.; Luo, Y.K. Changes in the Microbial Communities of Air-Packaged and Vacuum-Packaged Common Carp (Cyprinus carpio) Stored at 4 Degrees C. Food Microbiology 2015, 52, 197–204.
- Broekaert, K.; Heyndrickx, M.; Herman, L.; Devlieghere, F.; Vlaemynck, G. Molecular Identification of the Microbiota of Peeled and Unpeeled Brown Shrimp (Crangon crangon) during Storage on Ice and at 7.5 Degrees C. Food Microbiology 2013, 36(2), 123–134.
- Hozbor, M.C.; Saiz, A.I.; Yeannes, M.I.; Fritz, R. Microbiological Changes and Its Correlation with Quality Indices during Aerobic Iced Storage of Sea Salmon (Pseudopercis semifasciata). Lwt-Food Science and Technology 2006, 39(2), 99–104.
- Lalitha, K.V.; Surendran, P.K. Microbiological Changes in Farm Reared Freshwater Prawn (Macrobrachium rosenbergii De Man) in Ice. Food Control 2006, 17(10), 802–807.
- Ward, D.M.; Weller, R.; Bateson, M.M. 16S Rrna Sequences Reveal Numerous Uncultured Microorganisms in a Natural Community. Nature 1990, 345(6270), 63–65.
- Kim, H.J.; Kim, M.J.; Turner, T.L.; Kim, B.S.; Song, K.M.; Yi, S.H.; Lee, M.K. Pyrosequencing Analysis of Microbiota Reveals that Lactic Acid Bacteria are Dominant in Korean Flat Fish Fermented Food, Gajami-Sikhae. Bioscience Biotechnology and Biochemistry 2014, 78(9), 1611–1618.
- Madigan, T.L.; Bott, N.J.; Torok, V.A.; Percy, N.J.; Carragher, J.F.; Lopes, M.A.D.; Kiermeier, A. A Microbial Spoilage Profile of Half Shell Pacific Oysters (Crassostrea gigas) and Sydney Rock Oysters (Saccostrea glomerata). Food Microbiology 2014, 38, 219–227.
- Ojagh, S.M.; Rezaei, M.; Razavi, S.H.; Hosseini, S.M.H. Effect of Chitosan Coatings Enriched with Cinnamon Oil on the Quality of Refrigerated Rainbow Trout. Food Chemistry 2010, 120(1), 193–198.
- Hong, H.; Luo, Y.K.; Zhou, Z.Y.; Bao, Y.L.; Lu, H.; Shen, H.X. Effects of Different Freezing Treatments on the Biogenic Amine and Quality Changes of Bighead Carp (Aristichthys nobilis) Heads during Ice Storage. Food Chemistry 2013, 138(2–3), 1476–1482.
- Xiao, X.; Dong, Y.; Zhu, Y.; Cui, H.L. Bacterial Diversity Analysis of Zhenjiang Yao Meat during Refrigerated and Vacuum-Packed Storage by 454 Pyrosequencing. Current Microbiology 2013, 66(4), 398–405.
- Parlapani, F.F.; Boziaris, I.S. Monitoring of Spoilage and Determination of Microbial Communities Based on 16S Rrna Gene Sequence Analysis of Whole Sea Bream Stored at Various Temperatures. Lwt-Food Science and Technology 2016, 66, 553–559.
- Ozogul, F.; Gokbulut, C.; Ozogul, Y.; Ozyurt, G. Biogenic Amine Production and Nucleotide Ratios in Gutted Wild Sea Bass (Dicentrarchus labrax) Stored in Ice, Wrapped in Aluminium Foil and Wrapped in Cling Film at 4 Degrees C. Food Chemistry 2006, 98(1), 76–84.
- Bulushi, I.A.; Poole, S.; Deeth, H.; Dykes, G. Biogenic Amines in Fish: Roles in Intoxication, Spoilage, and Nitrosamine Formationa Review. Critical Reviews in Food Science and Nutrition 2009, 49(4), 369–377.
- Zhang, Y.M.; Qin, N.; Luo, Y.K.; Shen, H.X. Changes in Biogenic Amines and Atp-Related Compounds and Their Relation to Other Quality Changes in Common Carp (Cyprinus carpio Var. Jian) Stored at 20 and 0 Degrees C. Journal of Food Protection 2015, 78(9), 1699–1707.
- Krizek, M.; Vacha, F.; Vorlova, L.; Lukasova, J.; Cupakova, S. Biogenic Amines in Vacuum-Packed and Non-Vacuum-Packed Flesh of Carp (Cyprinus carpio) Stored at Different Temperatures. Food Chemistry 2004, 88(2), 185–191.
- Li, K.F.; Bao, Y.L.; Luo, Y.K.; Shen, H.X.; Shi, C. Formation of Biogenic Amines in Crucian Carp (Carassius auratus) during Storage in Ice and at 4 Degrees C. Journal of Food Protection 2012, 75(12), 2228–2233.
- Chytiri, S.; Chouliara, I.; Savvaidis, I.N.; Kontominas, M.G. Microbiological, Chemical and Sensory Assessment of Iced Whole and Filleted Aquacultured Rainbow Trout. Food Microbiology 2004, 21(2), 157–165.
- International Commission on Microbiological Specifications for Foods (ICMSF).
- Zare, D.; Ghazali, H.M. Assessing the Quality of Sardine Based on Biogenic Amines Using a Fuzzy Logic Model. Food Chemistry 2017, 221, 936–943.
- Hwang, C.C.; Lee, Y.C.; Huang, Y.R.; Lin, C.M.; Shiau, C.Y.; Hwang, D.F.; Tsai, Y.H. Biogenic Amines Content, Histamine-Forming Bacteria and Adulteration of Bonito in Tuna Candy Products. Food Control 2010, 21(6), 845–850.
- Bakar, J.; Yassoralipour, A.; Abu Bakar, F.; Rahman, R.A. Biogenic Amine Changes in Barramundi (Lates calcarifer) Slices Stored at 0 Degrees C and 4 Degrees C. Food Chemistry 2010, 119(2), 467–470.
- Zhu, Y.C.; Ma, L.Z.; Yang, H.; Xiao, Y.; Xiong, Y.L. Super-Chilling (−0.7 Degrees C) with High-Co2 Packaging Inhibits Biochemical Changes of Microbial Origin in Catfish (Clarias gariepinus) Muscle during Storage. Food Chemistry 2016, 206, 182–190.
- Tryfinopoulou, P.; Tsakalidou, E.; Nychas, G.J.E. Characterization of Pseudomonas Spp. Associated with Spoilage of Gilt-Head Sea Bream Stored under Various Conditions. Applied and Environmental Microbiology 2002, 68(1), 65–72.
- Koutsoumanis, K.; Lampropoulou, K.; Nychas, G.J.E. Biogenic Amines and Sensory Changes Associated with the Microbial Flora of Mediterranean Gilt-Head Sea Bream (Sparus aurata) Stored Aerobically at 0, 8, and 15 Degrees C. Journal of Food Protection 1999, 62(4), 398–402.
- Leisner, J.J.; Gram, L. Spoilage of Fish. In Encyclopedia of Food Microbiology; Robinson, R.K.; Batt, C.A.; Patel, P.D.; Eds.; San Diego, CA, USA: Academic Press, 1999; 813–820.
- Koutsoumanis, K.; Nychas, G.J.E. Application of a Systematic Experimental Procedure to Develop a Microbial Model for Rapid Fish Shelf Life Predictions. International Journal of Food Microbiology 2000, 60(2–3), 171–184.