ABSTRACT
Hepatocellular carcinoma (HCC) is one of the most lethal malignant tumors worldwide. Brahma-related gene 1 (BRG1), as a catalytic ATPase, is a major regulator of gene expression and is known to mutate and overexpress in HCC. The purpose of this study was to investigate the mechanism of action of BRG1 in HCC cells. In our study, BRG1 was silenced or overexpressed in human HCC cell lines. Transwell and wound healing assays were used to analyze cell invasiveness and migration. Mitochondrial membrane potential (MMP) and mitochondrial permeability transition pore (mPTP) detection were used to evaluate mitochondrial function in HCC cells. Colony formation and cell apoptosis assays were used to evaluate the effect of BRG1/TOMM40/ATP5A1 on HCC cell proliferation and apoptosis/death. Immunocytochemistry (ICC), immunofluorescence (IF) staining and western blot analysis were used to determine the effect of BRG1 on TOMM40, ATP5A1 pathway in HCC cells. As a result, knockdown of BRG1 significantly inhibited cell proliferation and invasion, promoted apoptosis in HCC cells, whereas BRG1 overexpression reversed the above effects. Overexpression of BRG1 can up-regulate MMP level, inhibit mPTP opening and activate TOMM40, ATP5A1 expression. Our results suggest that BRG1, as an oncogene, promotes HCC progression by regulating TOMM40 affecting mitochondrial function and ATP5A1 synthesis. Targeting BRG1 may represent a new and effective way to prevent HCC development.
Introduction
Hepatocellular carcinoma (HCC) is not only the predominant type of primary liver cancer, but also the leading cause of cancer-related death worldwide.Citation1–3 Globally, the incidence of HCC continues to rise, costing billions of dollars annually, which causes severe burden on the family and society.Citation2,Citation3 Due to the lack of specific biomarkers and classical symptoms, the vast majority of patients are diagnosed at advanced HCC.Citation4,Citation5 Although the curative treatment approaches such as liver transplantation and hepatectomy have yielded favorable outcomes in terms of overall survival, currently approved anti-HCC agents remain limited and face major challenges.Citation3,Citation6,Citation7 Therefore, it is imperative to understand the underlying molecular pathogenesis of HCC and to identify novel biomarkers or targets for the early diagnosis and treatment of HCC.
Genetic landscapes of HCC have revealed that HCC is characterized prominently by molecular and biological heterogeneity.Citation8,Citation9 Owing to the complex genetic heterogeneity of HCC disease, the antitumor efficacy of many targeted therapeutic drugs is limited.Citation9,Citation10 Thus, it is of great significance to identify an effective and precise treatment options for HCC.Citation11,Citation12,Citation13 Brahma-related gene 1 (BRG1), an ATPase component of SWI/SNF chromatin remodeling complex, is required for cell growth, differentiation and organ development to regulate a wide variety of physiological and pathological processes.Citation14,Citation15 BRG1 emerges as a coactivator and corepressor machinery that directly governs the transcription of genes, which is involved in cancer development.Citation16 However, the role of BRG1 in tumorigenesis remains largely controversial.Citation17 Accumulating studies show that loss or mutation of BRG1 functionally altered gene expression patterns of KRAS, TP53 and ATG16L1 in several cancers, including lung cancer and colorectal cancer.Citation18,Citation19 Additional studies suggest that overexpression of BRG1 was identified in various cancers, including pancreatic cancer, prostate cancer, glioblastoma and breast cancer.Citation14,Citation20–23 These data demonstrate the possibility that BRG1 serves either as an oncogene or tumor suppressor gene depending on the cellular and genetic milieu.
Recently, the public data evidenced by The Cancer Genome Atlas (TCGA) shows that increased BRG1 expression is positively associated with the severity of human HCC as well as metastasis.Citation24 In HCC, somatic heterozygous and mutations of BRG1 occur in ~ 3% of human HCC samples, resulting in overexpression.Citation24 Data from mouse models has shown that ablation of BRG1 blocked liver tumor formation in mice.Citation24 In addition, data from human HCC cell lines has demonstrated that BRG1 knockdown prevented cell proliferation, invasion and colony formation.Citation25 However, the underlying mechanism of BRG1 in HCC remain largely unknown at present. Studies have demonstrated a link between chromatin remodeling SWI/SNF components BRG1 and BRM and mitochondrial homeostasis.Citation26 Given the important role of TOMM40 and ATP5A1 in the regulatory mechanisms of mitochondrial dysfunction,Citation27,Citation28 we speculate that BRG1 may improve mitochondrial function by regulating TOMM40 and ATP5A1. In the study, we systematically analyzed the specific role of BRG1 in HCC cell lines. Our data support the hypothesis that BRG1, as an oncogene in HCC, promotes cell growth, invasion, and colony formation, and suppresses apoptosis through TOMM40/ATP5A1 pathway.
Material and methods
Cell culture
Human HCC cell lines HepG2 and HuH-7 were purchased from the Cell Libraries of Chinese Academy of Sciences (Shanghai, China). HepG2 and HuH-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco) in a 5% CO2 air atmosphere at 37°C.
Small RNA interference and transfection
Human BRG1 siRNA (#1, 5’-CCUCCGUGGUGAAGGUGUCTT-3’; 2#, 5’-GGUGAUCCACGUGGAGAGUTT-3’; 3#, 5’-GGAAUACCUCAAUAGCAUUTT-3’), TOMM40 siRNA (#1, 5’-GCAACAUACUACCACAAAGTT-3’; 2#, 5’-CUGGUUGGCAACGGUAACGTT-3’; 3#, 5’-GCACGCAACAUACUACCACTT-3’) and negative control (5’-CCUUUAAGGGUUCCCAACGTT-3’) were synthesized by the Sangon Biotech (Shanghai) Co., Ltd. Human BRG1 CDS was cloned into a pcDNA3.1 vector. Cells were planted into 6, 12, 24 or 96-well plates, and transfected with 100 nM siRNAs or indicated plasmids for 36 ~ 48 h using Lipo8000 reagent (Cat#C0533, Beyotime Biotech, Shanghai).
Western blotting
Whole cell lysates were extracted with RIPA lysis buffer containing a protease inhibitor (Cat#P0013C, Beyotime Biotech), and protein concentration was measured by a BCA kit (Cat#P0010S, Beyotime Biotech). Equal amounts of 30 μg protein were separated in an 8%~10% SDS-PAGE gel and transferred to a PVDF membrane (Cat#IPVH00010, Millipore). After blocking with 5% nonfat milk for 1 h at room temperature, the membranes were incubated with indicated primary antibodies: anti-rabbit BRG1 antibody (1:1000, 21634–1-AP, Proteintech, China), anti-rabbit TOMM40 antibody (1:1000, 18409–1-AP, Proteintech, China), anti-rabbit ATP5A1 (1:1000, 14676–1-AP, Proteintech, China) and anti-mouse β-actin antibody (1:2000, 66009–1-Ig, Proteintech, China) overnight at 4°C. Then, the blots were incubated with HRP-conjugated goat anti-rabbit or mouse IgG secondary antibody (1:6000, Cat# SA00001–2, Proteintech, China) for 1 h and were exposed in the SuperSignal West Pico Substrate (Cat#D3308–1, Beyotime Biotech). The gray value of all bands was determined by ImageJ software.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Reverse transcribe the extracted total RNA into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Thermo Fisher Scientific, Inc.), PowerUpTM SYBRTM Green Master Mix (Applied biosystems, Thermo Fisher Scientific, Inc.) was used to detect the mRNA expression level of TOMM40 and ATP5A1 after BRG1 overexpressed or knockdown. The primer was designed by SangonBiotech (Shanghai, China,https://www.sangon.com/).
Transwell invasion assay
The 500 μL/well complete DMEM containing 20% FBS was added to a 24-well plate, and a Transwell chamber (Corning) was placed in each well and treated with Matrigel (Cat#BME001, R&D systems) DMEM mixture at a ratio of 1:8. Then BRG1 overexpression or knockdown, TOMM40 knockdown and ATP5A1 knockdown cells were detached from culture substrate with 0.25% trypsin-EDTA (2508964, gibco, Canada), adjusted to 5 × 105 cells/mL and resuspended with 100 µL serum-free DMEM. The cell suspension was added to each chamber and cultured in an incubator. After incubation at 37°C for 24 h, cells were fixed with 4% paraformaldehyde (PFA) and stained with 0.1% crystal violet solution (Cat#G1062, Solarbio, China) for 30 min. Different fields of cells were randomly selected and photographed under a light microscope. Three microscope fields per condition were used for plotting the results.
Wound healing assay
Transfected HepG2 and HuH-7 cells (8 × 105 cells/well) were seeded in a 6-well plate and cultured in an incubator. When the cells reached 90%~100% confluence, the monolayer cells were wounded by scraping with a sterile pipette tip. Then, cells were exchanged in a serum-free medium and cultured in an incubator at 37°C. After washing in PBS, the cells were photographed at 0 h and 24 h by a light microscope, randomly selected fields were used for counting wound size.
Colony formation assay
The cells in each group (200 cells/well) were prepared in a single cell suspension, placed in a 6-well plate and cultured at 37°C in a 5% CO2 atmosphere. After incubation for 10–14 days, cells were gently washed by PBS, fixed with 4% PFA and stained with 0.1% crystal violet solution. The stained colonies were photographed and counted.
Cell apoptosis assay
Cell apoptosis assays were performed using Apoptosis and Necrosis Detection Kit with YO-PRO-1 and PI (Cat#C1075, Beyotime Biotech) in accordance with the manufacturer’s experiment procedures. Cells were seeded in a 96-well plate and treated with indicated plasmids. After transfection, the cells were washed in PBS for one time, and then 100 µL working buffer containing 1 µL YO-PRO-1 and 1 µL PI was added to the cells, followed by incubation in the dark environment at 37°C for 10 min. After treatment, the stained cells were photographed, counted and measured as the proportion of apoptotic cells.
TUNEL assay
Transfect adherent cells with TOMM40 siRNA and ATP5A1 siRNA and culture for 48 h, then fix the cells with 4% paraformaldehyde. Incubate at room temperature with PBS containing 0.3% Triton X-100 for 5 min. Incubate at room temperature with 0.3% H2O2 in PBS for 20 min to inactivate endogenous peroxidase. Incubate the samples sequentially with biotin labeling solution, Streptavidin HRP working solution, and DAB colorimetric solution (prepared according to the instructions of Colormetric TUNEL Apoptosis Assay Kit (C1098, Beyotime, China). After dehydration with gradient concentration alcohol, it can be sealed and stored for fluorescence detection.
Mitochondrial membrane potential (MMP) assay
Early apoptotic cells were evaluated using an MMP assay kit with JC-1 (Cat#C2006, Beyotime Biotech), which is a fluorescent probe to rapidly and sensitively detect changes in the membrane potential ∆Ψm of cells. Briefly, the cells in a 6-well plate were transfected and stained with 0.5 mM JC-1 in the dark environment at 37°C for 20 min. After washing three times with cold PBS, cells were detected under a Zeiss microscope (Germany). Red and green fluorescence intensity were analyzed by Image-Pro Plus 6.0 respectively. The ∆Ψm value for cell samples in each group was determined as the relative ratio of the red fluorescence intensity to the green fluorescence intensity.
mPTP detection
The cells were cultivated in 6-well plates and transfected. After the cells were washed with PBS for 1–2 times, reagent Calcein AM staining solution, Fluorescence quenching solution (CoCl2), and Ionomycin control were added to each well according to the instructions of Mitochondrial Permeability Transition Pore Assay Kit (C2009S, Beyotime, China), and the cells were incubated in a 37°C incubator for 45 min away from light. The cells were then incubated in a preheated fresh medium for another 30 min away from light. After incubation, the detection buffer can be added for observation under a fluorescence microscope.
Immunocytochemistry (ICC) and Immunofluorescence (IF)
For ICC staining, cells were seeded onto the coverslips treated with 0.1 mg/mL poly-L-lysine (Cat#A3890401, Gibco) and fixed with 4% paraformaldehyde for 10 min at room temperature, and then washed three times with ice-cold PBS. And cells were permeabilized with 0.1% Triton X-100 for 10 min. After washing in PBS three times for 5 min, cells were incubated with 1% BSA (Cat#ST2249, Beyotime Biotech), 0.1% Tween 20 in PBS for 30 min to block the unspecific binding of the antibodies, and then incubated with anti-rabbit BRG1 antibody (1:300) or anti-rabbit TOMM40 antibody (1:100) overnight at 4°C. Afterward, the cells were incubated with the secondary antibody (Cat# PV-9000, ZSGB-BIO, China) for 1 h and then stained with diaminobenzidin (DAB). Finally, cells were counterstained with hematoxylin (HE) again. The percentage of positive cells was counted by ImageJ software. For IF staining, cell samples were incubated with Dylight 488 fluorescent-labeled goat anti-rabbit secondary antibody (1:200, Cat# ZF-0512, ZSGB-BIO, China), and then nuclei were stained with 5 μg/mL DAPI (Cat# ZLI-9557, ZSGB-BIO, China). Cell samples were observed under a Zeiss microscope, and then the fluorescence intensity was analyzed by Image-Pro Plus 6.0 software.
Statistical analysis
All representative data are shown as the mean ± SEM. Statistical analysis was conducted using GraphPad Prism 9.0 software. Differences between groups or multiple groups were analyzed using a two-tailed Student’s t-test or One-way ANOVA test. Statistical significance was considered at p < .05.
Results
Effects of BRG1 knock-down and overexpression on the invasion and migration of HCC cells
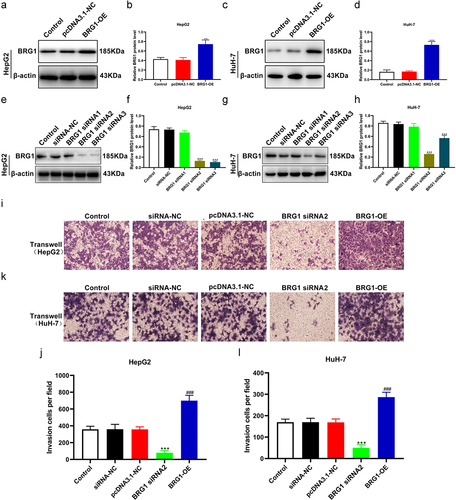
Both TCGA liver hepatocellular carcinoma (LIHC) and Fudan datasets revealed that BRG1 is overexpressed in human HCC specimens.Citation24 Therefore, we next systematically investigated the specific role of BRG1 in HCC cell lines. We overexpressed BRG1 using pcDNA3.1-BRG1 and knocked down BRG1 in HepG2 and HuH-7 cells using specific siRNAs. As shown in (), pcDNA3.1-BRG1 significantly upregulated the expression of BRG1 in HepG2 and HuH-7 cells compared with pcDNA3.1 vector (), while BRG siRNA#2 or #3 could markedly decrease the expression in both HepG2 and HuH-7 cells compared with negative control (NC) siRNA (). In the subsequent in vitro experiments, we chose siRNA#2 as a knockdown tool for BRG1. Transwell assay showed that overexpression of BRG1 significantly promoted the invasion of HepG2 and HuH-7 cells compared with pcDNA3.1 vector (), whereas siRNA#2-mediated BRG1 knockdown obviously prevented cell invasion compared with NC siRNA ()). These results suggest that BRG1 is positively correlated with HCC progression.
Figure 1. BRG1 promotes HCC cell invasion. (a) HepG2 cells were transfected with pcDNA3.1 or BRG1 overexpression plasmid for 48 h, and then cell lysates were performed using western blot assay. (b) The gray blots were analyzed with ImageJ software (n = 3). **p < .01 vs pcDNA3.1-NC. (c) HuH-7 cells transfected with BRG1 plasmid were analyzed using western blot assay. (d) The gray blots were analyzed with ImageJ software (n = 3). ***p < .001 vs pcDNA3.1-NC. (e) The BRG1 siRNAs were designed and tranfected for 36 h into HepG2 cells, and then accessed using western blot assay. (f) The gray blots were analyzed with ImageJ software (n = 3). ***p < .001 vs siRNA-NC. (g) The knockdown effect of siRNA-BRG1 in HuH-7 cells were evaluate by western blot assay. (h) The gray blots were analyzed with ImageJ software (n = 3). ***p < .001 vs siRNA-NC. (i) Transwell assay was used to detect the invasion ability after BRG1 knockdown or overexpression in HepG2 cells. Scale bar, 50 μm. (j) Cells of different fields were randomly selected and counted (n = 3). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC. (k) The invasion ability after BRG1 knockdown or overexpression in HuH-7 cells was detected by transwell assay. Scale bar, 50 μm. (l) Cells of different fields were randomly selected and counted (n = 3). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC.
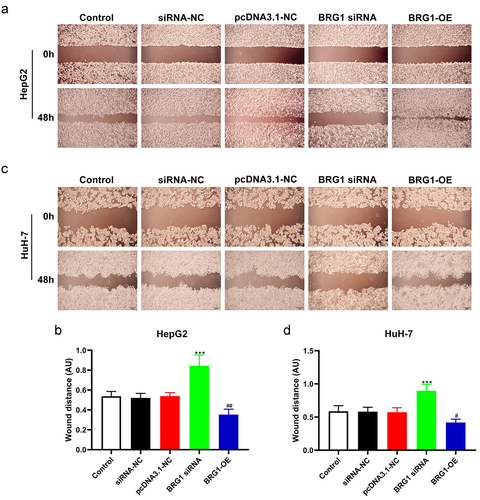
To further investigate the role of BRG1 on the migration of HCC cells, we evaluated the migration ability of HCC cells by wound healing assay (). The wound healing assay showed that overexpression of BRG1 in HepG2 and HuH-7 cells enhanced cell migration at the edge of the wound compared with the control (), while BRG1 siRNA#2 significantly inhibited cell migration and shortened the migration distance of HCC cells at 48 h compared with NC siRNA (). Based on these findings, BRG1 servers as an oncogene in HCC and may directly or indirectly contribute to HCC development.
Figure 2. BRG1 affects HCC cell migration. (a) Wound healing assay was used to evaluate the migration ability at 48 h after BRG1 knockdown or overexpression in HepG2 cells. Scale bar, 200 μm. (b) Different fields of cells were randomly selected, and then wound size was calculated (n = 3). ***p < .001 vs siRNA-NC, ##p < .01 vs pcDNA3.1-NC. (c) The migration ability after BRG1 knockdown or overexpression in HuH-7 cells was evaluated by wound healing assay. Scale bar, 200 μm. (d) Different fields of cells were randomly selected, and then wound size was calculated (n = 3). ***p < .001 vs siRNA-NC, #p < .05 vs pcDNA3.1-NC.
Effects of BRG1 knock-down and overexpression on the apoptosis and proliferation of HCC cells
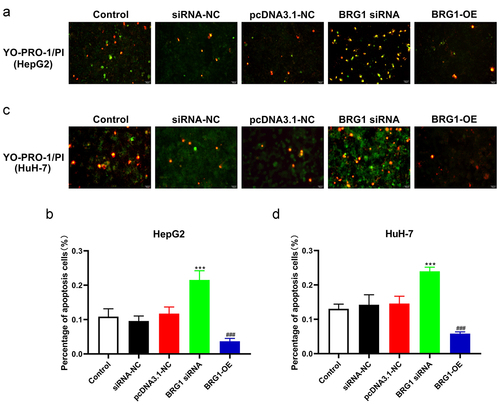
To further investigate the apoptotic effect of BRG1 in HCC cells, we then performed a battery of experiments in vitro. YO-PRO-1, also known as oxazole yellow, is commonly used to detect cell apoptosis and shows specific binding to DNA in apoptotic cells. However, the necrotic cells can also be stained with YO-PRO-1, which in combination with PI, namely propidium iodide, is responsible for determining specific necrotic cells to effectively identify apoptosis (). Green fluorescent staining indicated that compared with negative control, BRG1 siRNA#2 significantly promoted the apoptosis of HepG2 and HuH-7 cells (), whereas overexpression of BRG1 obviously inhibited cell apoptosis compared with the control ().
Figure 3. BRG1 inhibits HCC cell apoptosis. (a) HepG2 cells were transfected with BRG1 siRNA or overexpression plasmid, respectively, and then stained with both YO-PRO-1 and PI dye. Scale bar, 50 μm. (b) The proportion of green-labeled apoptotic cells were measured accroding to randomly selected cell fields (n = 5). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC. (c) HuH-7 cells transfected with BRG1 siRNA or overexpression plasmid were stained with both YO-PRO-1 and PI dye, respectively. Scale bar, 50 μm. (d) The proportion of apoptotic cells were measured (n = 5). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC.
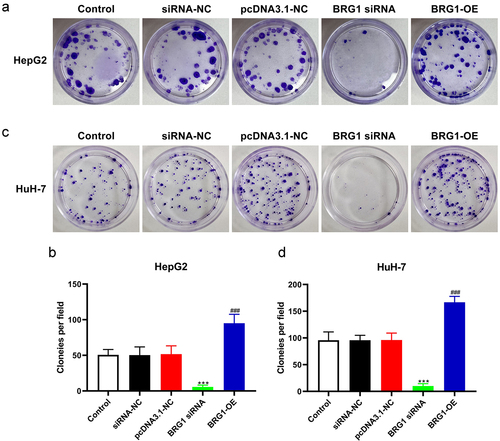
We next investigate the proliferation ability of BRG1 in HCC cells by using colony formation assay (). After HCC cells were transfected with BRG1 siRNA, the growth rate in BRG1 knockdown group was significantly inhibited compared with the negative control group (). Conversely, the growth rate in BRG1 overexpression group was markedly increased compared with the control group in both HepG2 and HuH-7 cells (). Collectively, these findings suggest that BRG1 promotes HCC cell proliferation and inhibits HCC cell apoptosis.
Figure 4. BRG1 increases HCC cell colony formation. (a) Colony formation assay was used to evaluate the colony formation ability after BRG1 knockdown or overexpression in HpG2 cells. (b) The stained colonies were photographed and counted (n = 5). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC. (c) The colony formation ability after BRG1 knockdown or overexpression in HuH-7 cells was examined. (d) The stained colonies were photographed and counted (n = 5). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC.
Effects of BRG1 knock-down and overexpression on the mitochondrial function of HCC cells
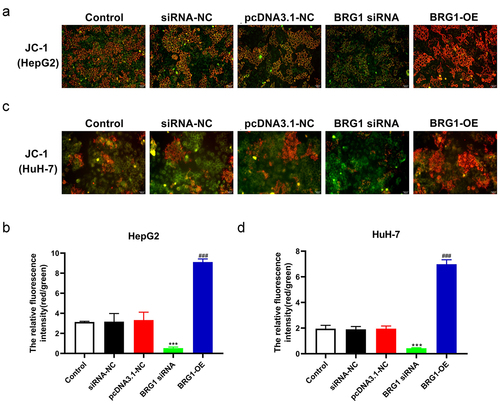
JC-1, an ideal fluorescent probe, is widely used to detect the MMP ∆Ψm, the decrease of which is a hallmark event in the early stage of apoptosis.Citation29 MPP assay showed that compared with negative control, BRG1 siRNA#2 markedly abolished the transition of JC-1 from red to green fluorescence in HCC cells (), while BRG1 overexpression increased the relative ratio of red to green fluorescence compared with the control ().
Figure 5. BRG1 alters the membrane potential ΔΨm of HCC cells. (a) MPP assay was used to analyze membrane potential ΔΨm, and then HepG2 cells were stained with JC-1 probe after BRG1 knockdown or overexpression. Scale bar, 50 μm. (b) The relative ratio of the red fluorescence intensity to the green fluorescence intensity was determined (n = 5). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC. (c) HuH-7 cells with treatment were stained with JC-1 probe. Scale bar, 50 μm. (d) The relative fluorescence intensity was determined by ImageJ software (n = 5). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC.
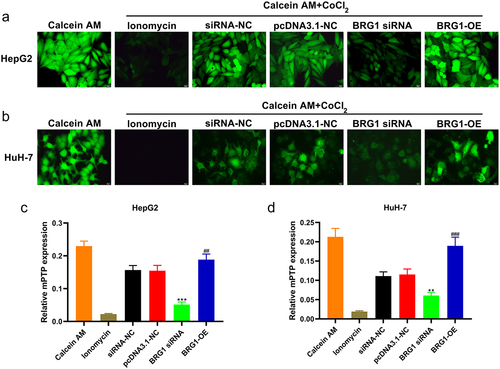
The decrease of mitochondrial membrane potential also leads to the opening of Mitochondrial Permeability Transition Pore (mPTP). In mPTP detection assay, compared with siRNA-NC group, BRG1 siRNA#2 reduced mitochondrial membrane potential and promoted the opening of mPTP (), and overexpression of BRG1 significantly inhibited the opening of mPTP (). Collectively, these findings suggest that BRG1 could promote HCC cell mitochondrial function.
Figure 6. BRG1 regulates the opening of mitochondrial permeability transition pore of HCC cells. (A) HepG2 cells were stained with Calcein AM after BRG1 knockdown or overexpression. Scale bar, 20 μm. (B) Fluorescence intensity of different treatment groups was determined (n = 5). ***p < .001 vs siRNA-NC, ##p < .01 vs pcDNA3.1-NC. (C) HuH-7 cells with treatment were stained with Calcein AM. Scale bar, 20 μm. (D) Fluorescence intensity of different treatment groups was determined (n = 5). **p < .01 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC.
BRG1 regulates HCC development through TOMM40/ATP5A1 pathway
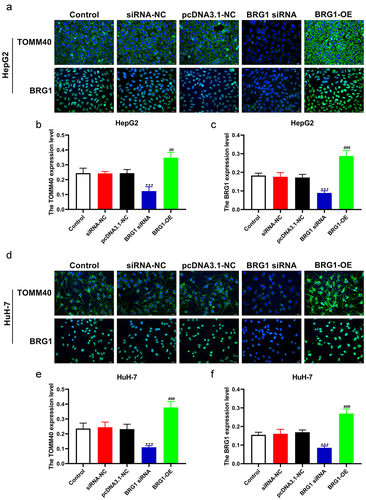
Multiple studies indicated that TOMM40 variants were detected in patients with age-related neurodegenerative disorders.Citation30,Citation31 However, the association between either TOMM40 and HCC or TOMM40 and BRG1 has not been elucidated. To investigate the correlation between TOMM40 expression and BRG1 in HCC cells, we performed ICC and IF analysis (). ICC results revealed that the expression of TOMM40 was significantly decreased in BRG1 knockdown HCC cells compared with negative control (), whereas TOMM40 expression was markedly increased in BRG1 overexpression HCC cells (). Similarly, IF staining also indicated that TOMM40 expression was significantly downregulated in siRNA-mediated BRG1 knockdown HCC cells compared with negative control (), while TOMM40 expression was markedly increased in BRG1 overexpression cells (). These results suggest that TOMM40 can be a candidate target of BRG1.
Figure 7. BRG1 induces the expession of TOMM40 in HCC cells. (a) HepG2 cells were stained with TOMM40 and BRG1 antibody after BRG1 knockdown or overexpression. (b and c) the percentage of positive cells was analyzed by ImageJ software (n = 10). **p < .01, ***p < .001 vs siRNA-NC, ##p < .01, ###p < .001 vs pcDNA3.1-NC. (d) HuH-7 cells were stained with TOMM40 and BRG1 antibody after BRG1 knockdown or overexpression. (e and f) the percentage of positive cells was analyzed by ImageJ software (n = 10). ***p < .001 vs siRNA-NC, ##p < .01, ###p < .001 vs pcDNA3.1-NC.
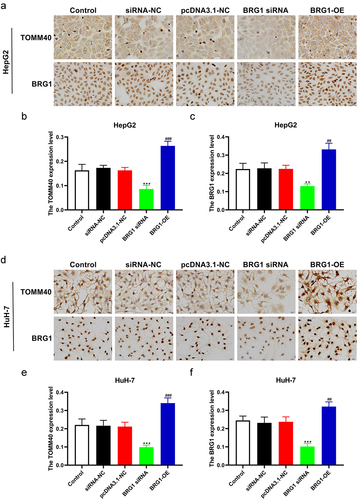
Figure 8. BRG1 induces the expession of TOMM40 in HCC cells. (a) HepG2 cells were immunostained with TOMM40 and BRG1 antibody after BRG1 knockdown or overexpression. Scale bar, 20 μm. (b and c) the relative fluorescence intensity was analyzed by ImageJ software (n = 10). ***p < .001 vs siRNA-NC, ##p < .01, ###p < .001 vs pcDNA3.1-NC. (d) HuH-7 cells were immunostained with TOMM40 and BRG1 antibody after BRG1 knockdown or overexpression. Scale bar, 20 μm. (e and f) the relative fluorescence intensity was analyzed by Image J software (n = 10). ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC.
In addition, ATP synthase alpha 1 subunit (ATP5A1), also named as ATP5F1A, has been reported that also plays oncogenic or tumor-suppressing roles in several malignancies, such as glioblastoma, colon adenocarcinoma, and renal cell carcinoma (RCC).Citation32–34 According to the available data, we then provide a scientific hypothesis whether BRG1/TOMM40 signaling is involved in regulating gene expression of ATP5A1 in HCC cells. To further elucidate the correlation between ATP5A1 and BRG1/TOMM40 pathway in HCC cells, we detected ATP5A1 expression in BRG1 overexpression or knockdown cells using western blotting assay (). Results showed that the expressions of TOMM40 and ATP5A1 was significantly upregulated in BRG1 overexpression cells compared with the control (), whereas ATP5A1 expression was obviously downregulated in BRG1 knockdown cells compared with negative control (). The results of RT qPCR showed that the mRNA expression levels of TOMM40 and ATP5A1 were similarly regulated by BRG1 (). Moreover, we designed TOMM40 siRNAs () and ATP5A1 siRNA to confirm whether ATP5A1 is the downstream effector of BRG1/TOMM40 axis and the effects of TOMM40 and ATP5A1 on cell growth, apoptosis and invasion. As expected, TOMM40 knockdown markedly resulted in a reduction of ATP5A1 expression in HCC cells but cannot affect the expression of BRG1, and ATP5A1 knockdown had no influence on neither BRG1 nor TOMM40 (). Knocking out both TOMM40 and ATP5A1 can significantly inhibit the proliferation and migration of liver cancer cells, while promoting HCC cell apoptosis (). These data suggest that ATP5A1 may serve as a potential downstream effector of BRG1/TOMM40 pathway and both play important roles in cell growth, apoptosis and invasion.
Figure 9. BRG1 activates TOMM40/ATP5A1 pathway in HCC cells. (a) The expressions of BRG1, TOMM40 and ATP5A1 were analyzed using Western blotting after BRG1 knockdown or overexpression in HepG2 cells. (b-d) the relative intensities of BRG1, TOMM40 and ATP5A1 were determined using ImageJ software (n = 3). **p < .01, ***p < .001 vs siRNA-NC, ##p < .01, ###p < .001 vs pcDNA3.1-NC. (e) The expressions of BRG1, TOMM40 and ATP5A1 were analyzed using Western blotting after BRG1 knockdown or overexpression in HuH-7 cells. (f-h) the relative intensity were determined using ImageJ software (n = 3). **p < .01, ***p < .001 vs siRNA-NC, ###p < .001 vs pcDNA3.1-NC. (i-l) the mRNA expressions level of BRG1, TOMM40 and ATP5A1 were analyzed using RT-qPCR.(n = 3). ##p < .01, ###p < .001 vs siRNA-NC, ***p < .001 vs pcDNA3.1-NC.
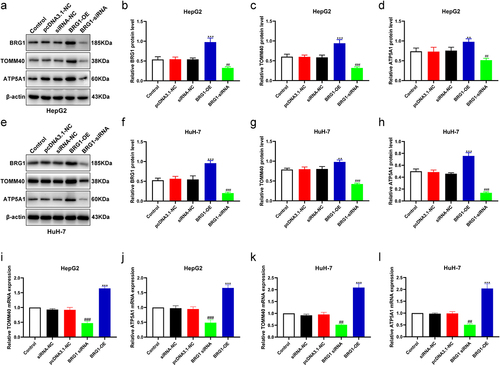
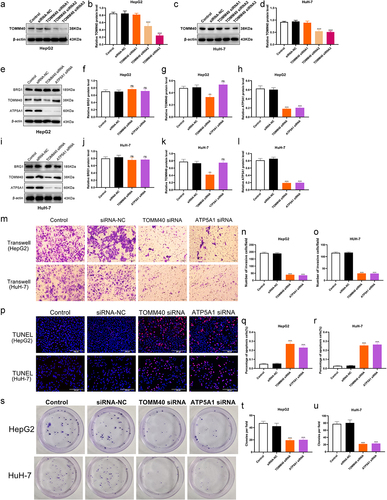
Figure 10. TOMM40 and ATP5A1 on cell growth, apoptosis and invasion. (a, c) HCC cells were transfected with TOMM40 siRnas, and then cell lysates were performed using Western blotting. (b, d) the relative expression of TOMM40 were determined by ImageJ software (n = 3). ***p < .001 vs siRNA-NC. (e, i) the expressions of BRG1, TOMM40 and ATP5A1 were analyzed using Western blotting after TOMM40 and ATP5A1 knockdown in HepG2 and HuH-7 cells. (f-h, j-l) the relative intensity of blots was determined using ImageJ software (n = 3). ns: not significant, **p < .01, ***p < .001 vs siRNA-NC. (m-o) Transwell assay was used to detect the invasion ability after TOMM40 or ATP5A1 knockdown in HCC cells. Scale bar, 50 μm. (n = 3). (p-R) TUNEL assay was used to detect the proportion of apoptotic cells after TOMM40 or ATP5A1 knockdown in HCC cells. Scale bar, 200 μm. (n = 3). (s-u) Colony formation assay was used to evaluate the colony formation ability after TOMM40 or ATP5A1 knockdown in HCC cells. (n = 3). ***p < .001 vs siRNA-NC.
Discussion
There is growing evidence for the importance of the SWI/SNF complex during tumorigenesis. A number of genome-wide sequencing studies have revealed a remarkably high prevalence of mutations in the SWI/SNF complex in nearly 25% of a wide variety of human cancers.Citation35–37 Functional genomic studies have indicated that dysfunction of mammalian SWI/SNF enzymes leads to some constitutively overexpressed genes, which in turn drive cancer development.Citation37,Citation38 The SWI/SNF complex consists of the BRG1-associated factor (BAF) and polybromo BRG1-associated factor (PBAF).Citation38 BRG1, encoded by the SMARCA4 gene, is a catalytic subunit of SWI/SNF chromatin remodeling factor.Citation24,Citation38 The discovery that loss of BRG1 activity is a major driver for developing aggressive tumors,Citation18,Citation19,Citation39 while BRG1 are also abnormally amplified in many cancers,Citation14,Citation20,Citation24 highlighting its oncogenic potential, the roles of which in cancer appears to be strongly dependent on the type of cancer.
Nevertheless, an increasing number of studies have revealed the positive role of BRG1 in HCC initiation and progression.Citation24,Citation25 TCGA and other datasets exhibited a statistical evidence for a mutual attraction pattern between BRG1 and HCC.Citation24 Interestingly, KEGG pathway analysis suggested that BRG1-correlated co-expressed genes were significantly enriched in HCC pathway.Citation24 More importantly, TCGA LIHC studies showed that dramatically amplification of BRG1 was positively correlated with poor HCC patient survival.Citation24 In addition, BRG1 deficiency in endothelial cells attenuates liver fibrosis in mice.Citation40 In the study, we explore the biological role of BRG1 in HCC cell lines, and then confirmed that overexpression of BRG1 can significantly promote HepG2 and HuH-7 cell invasion, migration and proliferation capabilities. Conversely, knockdown of BRG1 played a role in inhibiting these effects in HCC cells. Consistent with our data, BRG1 deleption in mouse hepatocytes or HCC cell lines prevented hepatocellular carcinoma (HCC) formation and growth.Citation24,Citation25 In addition, we first detected the apoptotic effect of BRG1 in HCC cells, and then confirmed that BRG1 knockdown can activate apoptotic procedure in HCC cells, while BRG1 overexpression significantly abolished apoptotic pathway, suggesting a possible antiapoptotic role of BRG1 in HCC. Recent studies have shown that mitochondria are involved in the regulation of apoptosis pathway, which is related to the decrease of mitochondrial membrane potential and the opening of mitochondrial permeability transition pore.Citation41–43 Our results also showed that BRG1 knockdown decreased MMP in HCC cells and was accompanied by the opening of mPTP. Importantly, BRG1 has been shown to own both and tumor-suppressor and carcinogenic roles in many cancers, nevertheless our study and previous reports identified BRG1 as an oncogene affecting HCC development.Citation24,Citation25
TOMM40, localized in the outer membrane of the mitochondria, is essential for the formation of a translocase of the outer mitochondrial membrane (TOMM) complex, which is a major biological contributor to mitochondrial dysfunction.Citation30 Previous data demonstrate that a TOMM40 variable polymorphism has been implicated in neurodegenerative disorders.Citation30,Citation31 However, the detailed molecular pathology of how BRG1 and TOMM40 in HCC remains largely unknown. We investigated the association between BRG1 and TOMM40 in HCC cells using immunostaining and western blot assays. We found that there is a statistically significant positive correlation between BRG1 and TOMM40 in HCC cells, suggesting that BRG1/TOMM40 pathway may exhibit specific roles in HCC progression. In addition, our data further investigated the association between the activation of BRG1/TOMM40 pathway and ATP5A1 expession, and we found that the overexpression of either BRG1 or TOMM40 can significantly increase ATP5A1 expression in HCC cells, whereas the knockdown of them obviously suppressed ATP5A1 expression. Mounting evidence indicates that ATP5A1, a subunit of mitochondrial ATP synthase, has been recently reported to be linked with several malignancies, nevertheless it was recognized as an either oncogene or tumor suppression gene in different cancer types.Citation32–34 Therefore, the role of ATP5A1 in antitumor and tumorigenesis remains largely controversial.Citation34 Based on these data, our study highlights the functional importance of BRG1/TOMM40/ATP5A1 pathway in HCC formation and progression. However, the cancer-related roles of BRG1 seem to be distinct in different cancer types.
Supplemental Material
Download MS Word (14.7 KB)Supplementary Figure.jpg
Download JPEG Image (2.6 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2375440
Additional information
Funding
References
- Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–91.e471. doi:10.1053/j.gastro.2018.08.065.
- Villanueva A, Longo DL. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–15. doi:10.1056/NEJMra1713263.
- Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet (Lond, Engl). 2022;400(10360):1345–62. doi:10.1016/S0140-6736(22)01200-4.
- Frenette C. Advances in hepatocellular carcinoma. Clin Liver Dis. 2020;24(4):xiii–xiv. doi:10.1016/j.cld.2020.08.014.
- Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6):1370. doi:10.3390/cells9061370.
- Galle PR, Dufour JF, Peck-Radosavljevic M, Trojan J, Vogel A. Systemic therapy of advanced hepatocellular carcinoma. Future Oncol (Lond, Engl). 2021;17(10):1237–51. doi:10.2217/fon-2020-0758.
- Sugawara Y, Hibi T. Surgical treatment of hepatocellular carcinoma. Bioscience trends. Biosci Trends. 2021;15(3):138–41. doi:10.5582/bst.2021.01094.
- Shibata T. Genomic landscape of hepatocarcinogenesis. J Hum Genet. 2021;66(9):845–51. doi:10.1038/s10038-021-00928-8.
- Takeda H, Takai A, Eso Y, Takahashi K, Marusawa H, Seno H. Genetic landscape of multistep hepatocarcinogenesis. Cancers. 2022;14(3):568. doi:10.3390/cancers14030568.
- Javan H, Dayyani F, Abi-Jaoudeh N. Therapy in advanced hepatocellular carcinoma. Semin Intervent Radiol. 2020;37(5):466–74. doi:10.1055/s-0040-1719187.
- Hsu CL, Ou DL, Bai LY, Chen C-W, Lin L, Huang S-F, Cheng A-L, Jeng Y-M, Hsu C. Exploring markers of exhausted CD8 T Cells to predict response to immune checkpoint inhibitor therapy for hepatocellular carcinoma. Liver Cancer. 2021;10(4):346–59. doi:10.1159/000515305.
- Singh A, Beechinor RJ, Huynh JC, Li D, Dayyani F, Valerin JB, Hendifar A, Gong J, Cho M. Immunotherapy updates in advanced hepatocellular carcinoma. Cancers. 2021;13(9):2164. doi:10.3390/cancers13092164.
- Xing R, Gao J, Cui Q, Wang Q. Strategies to improve the antitumor effect of immunotherapy for hepatocellular carcinoma. Front Immunol. 2021;12:783236. doi:10.3389/fimmu.2021.783236.
- Giles KA, Gould CM, Achinger-Kawecka J, Page SG, Kafer GR, Rogers S, Luu P-L, Cesare AJ, Clark SJ, Taberlay PC, et al. BRG1 knockdown inhibits proliferation through multiple cellular pathways in prostate cancer. Clin Epigenetics. 2021;13(1):37. doi:10.1186/s13148-021-01023-7.
- Sobczak M, Strachowska M, Robaszkiewicz A. Contribution of BRG1-dependent SWI/SNF complexes to determining the phenotype of cancer cell. Postepy Biochemii. 2020;66(1):10–18. doi:10.18388/pb.2020_312.
- Mardinian K, Adashek JJ, Botta GP, Kato S, Kurzrock R. SMARCA4: implications of an altered chromatin-remodeling gene for cancer development and therapy. Mol Cancer Ther. 2021;20(12):2341–51. doi:10.1158/1535-7163.MCT-21-0433.
- Wu Q, Lian JB, Stein JL, Stein GS, Nickerson JA, Imbalzano AN. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics. 2017;9(6):919–31. doi:10.2217/epi-2017-0034.
- Dagogo-Jack I, Schrock AB, Kem M, Jessop N, Lee J, Ali SM, Ross JS, Lennerz JK, Shaw AT, Mino-Kenudson M, et al. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. J Thorac Oncol. 2020;15(5):766–76. doi:10.1016/j.jtho.2020.01.002.
- Liu M, Sun T, Li N, Peng J, Fu D, Li W, Li L, Gao W-Q. BRG1 attenuates colonic inflammation and tumorigenesis through autophagy-dependent oxidative stress sequestration. Nat Commun. 2019;10(1):4614. doi:10.1038/s41467-019-12573-z.
- Ding Y, Li N, Dong B, Guo W, Wei H, Chen Q, Yuan H, Han Y, Chang H, Kan S, et al. Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. J Clin Invest. 2019;129(2):759–73. doi:10.1172/JCI123557.
- Sun L, Yuan Y, Chen J, Ma C, Xu Y. Brahma related gene 1 (BRG1) regulates breast cancer cell migration and invasion by activating MUC1 transcription. Biochem Bioph Res Co. 2019;511(3):536–43. doi:10.1016/j.bbrc.2019.02.088.
- Tsuda M, Fukuda A, Roy N, Hiramatsu Y, Leonhardt L, Kakiuchi N, Hoyer K, Ogawa S, Goto N, Ikuta K, et al. The BRG1/SOX9 axis is critical for acinar cell–derived pancreatic tumorigenesis. J Clin Invest. 2018;128(8):3475–89. doi:10.1172/JCI94287.
- Wang Y, Yang CH, Schultz AP, Sims MM, Miller DD, Pfeffer LM. Brahma-Related Gene-1 (BRG1) promotes the malignant phenotype of glioblastoma cells. J Cell Mol Med. 2021;25(6):2956–66. doi:10.1111/jcmm.16330.
- Wang P, Song X, Cao D, Cui K, Wang J, Utpatel K, Shang R, Wang H, Che L, Evert M, et al. Oncogene-dependent function of BRG1 in hepatocarcinogenesis. Cell Death Dis. 2020;11(2):91. doi:10.1038/s41419-020-2289-3.
- Kaufmann B, Wang B, Zhong S, Laschinger M, Patil P, Lu M, Assfalg V, Cheng Z, Friess H, Hüser N, et al. BRG1 promotes hepatocarcinogenesis by regulating proliferation and invasiveness. PLoS One. 2017;12(7):e0180225. doi:10.1371/journal.pone.0180225.
- Bultman SJ, Holley DW, G GDR, Pizzo SV, Sidorova TN, Murray KT, Jensen BC, Wang Z, Bevilacqua A, Chen X, et al. BRG1 and BRM SWI/SNF ATPases redundantly maintain cardiomyocyte homeostasis by regulating cardiomyocyte mitophagy and mitochondrial dynamics in vivo. Cardiovasc Pathol. 2016;25(3):258–69. doi:10.1016/j.carpath.2016.02.004.
- Tang Z, Takahashi Y, Wang HG. ATG2 regulation of phagophore expansion at mitochondria-associated ER membranes. Autophagy. 2019;15(12):2165–66. doi:10.1080/15548627.2019.1666594.
- Liu X, Li M, Chen Z, Yu Y, Shi H, Yu Y, Wang Y, Chen R, Ge J. Mitochondrial calpain-1 activates NLRP3 inflammasome by cleaving ATP5A1 and inducing mitochondrial ROS in CVB3-induced myocarditis. Basic Res Cardiol. 2022;117(1):40. doi:10.1007/s00395-022-00948-1.
- Komatsu S, Nomiyama T, Numata T, Kawanami T, Hamaguchi Y, Iwaya C, Horikawa T, Fujimura-Tanaka Y, Hamanoue N, Motonaga R, et al. SGLT2 inhibitor ipragliflozin attenuates breast cancer cell proliferation. Endocr J. 2020;67(1):99–106. doi:10.1507/endocrj.EJ19-0428.
- Lee EG, Chen S, Leong L, Tulloch J, Yu CE. TOMM40 RNA transcription in alzheimer’s disease brain and its implication in mitochondrial dysfunction. Genes. 2021;12(6):871. doi:10.3390/genes12060871.
- Zhu Z, Yang Y, Xiao Z, Zhao Q, Wu W, Liang X, Luo J, Cao Y, Shao M, Guo Q, et al. TOMM40 and APOE variants synergistically increase the risk of Alzheimer’s disease in a Chinese population. Aging Clin Exp Res. 2021;33(6):1667–75. doi:10.1007/s40520-020-01661-6.
- Xu G, Li JY. ATP5A1 and ATP5B are highly expressed in glioblastoma tumor cells and endothelial cells of microvascular proliferation. J Neurooncol. 2016;126(3):405–13. doi:10.1007/s11060-015-1984-x.
- Yuan L, Chen L, Qian K, Wang G, Lu M, Qian G, Cao X, Jiang W, Xiao Y, Wang X, et al. A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by co‑expression analysis. Oncol Rep. 2018;39(2):525–36. doi:10.3892/or.2017.6132.
- Zhang G, Zhong J, Lin L, Liu Z. Loss of ATP5A1 enhances proliferation and predicts poor prognosis of colon adenocarcinoma. Pathol Res Pract. 2022;230:153679. doi:10.1016/j.prp.2021.153679.
- Fukumoto T, Magno E, Zhang R. SWI/SNF complexes in ovarian cancer: Mechanistic Insights and therapeutic implications. Mol Cancer Res. 2018;16(12):1819–25. doi:10.1158/1541-7786.MCR-18-0368.
- Mittal P, Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17(7):435–48. doi:10.1038/s41571-020-0357-3.
- Xiao L, Parolia A, Qiao Y, Bawa P, Eyunni S, Mannan R, Carson SE, Chang Y, Wang X, Zhang Y, et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature. 2022;601(7893):434–39. doi:10.1038/s41586-021-04246-z.
- Wanior M, Krämer A, Knapp S, Joerger AC. Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. Oncogene. 2021;40(21):3637–54. doi:10.1038/s41388-021-01781-x.
- Li Z, Xia J, Fang M, Xu Y. Epigenetic regulation of lung cancer cell proliferation and migration by the chromatin remodeling protein BRG1. Oncogenesis. 2019;8(11):66. doi:10.1038/s41389-019-0174-7.
- Shao J, Xu Y, Fang M. BRG1 deficiency in endothelial cells alleviates thioacetamide induced liver fibrosis in mice. Biochem Bioph Res Co. 2020;521(1):212–19. doi:10.1016/j.bbrc.2019.10.109.
- Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends in cell biology. Trends Cell Biol. 2008;18(4):165–73. doi:10.1016/j.tcb.2008.01.006.
- Lim SY, Davidson SM, Hausenloy DJ, Yellon DM. Preconditioning and postconditioning: the essential role of the mitochondrial permeability transition pore. Cardiovasc Res. 2007;75(3):530–35. doi:10.1016/j.cardiores.2007.04.022.
- Armstrong J. The role of the mitochondrial permeability transition in cell death. Mitochondrion. 2006;6(5):225–34. doi:10.1016/j.mito.2006.07.006.
