Abstract
Laser Microdissection (LM) is a technology that allows the rapid procurement of selected cell populations from a section of heterogeneous tissues in a manner conducive to the extraction of DNA, RNA, proteins and even metabolites. In the past few years, it has also been applied to plant biology in order to study gene expression in plant-nematode and plant-microbe interactions. LM represents a powerful tool since cells associated with a particular infection stage can be visualized under the microscope and harvested. Therefore, verification of the response of the plant during the progression of the colonization can be performed in different cell types. Applications of LM to study the interaction between the plant and both pathogenic and symbiotic organisms (i.e. nematode and fungi, respectively) are explored in this review.
Introduction
Plant growth and development take place in a complex cellular environment (Day et al. Citation2005), where interactions with symbiotic and pathogenic organisms play important roles. A goal in biology is to identify genes that function in the cells that are specifically involved in these interactions. Several types of methods have been developed to demonstrate gene expression in various cell or tissue types. In situ hybridization and experiments with transgenic plants that harbor the β-glucuronidase (Jefferson et al. Citation1987) or enhanced green fluorescent protein (eGFP) (Haseloff et al. Citation1997) have been performed for many years. In these experiments, the molecular tag is linked to gene promoter elements or to the genes, themselves. This has allowed the visualization of localized expression in specific cell-types. However, efficient methods to isolate homogeneous populations of specific cell types are needed to fully understand the contribution of individual cells to the biology of an organism. By isolating homogeneous populations of cells, researchers could avoid diluting gene expression signals as a result of the presence of unwanted cells in their samples. Efficient methods for routinely isolating homogeneous cell populations are now available: these allow the analysis of thousands of transcripts from a wide range of cell or tissue types to understand the contribution of individual cells to the biology of an organism. Cell isolation can be achieved by enzymatic or physical means. Enzymatic means would rely on enzymes to digest cell walls, leaving protoplasts that could be sorted, typically by the use of fluorescent markers (Birnbaum et al. Citation2003). Physical means would be Laser Microdissection (LM).
LM is a technology that permits rapid procurement of selected cell populations from a histological tissue section that is composed of heterogeneous cells (Emmert-Buck et al. Citation1996). This procedure is performed in a manner that is conducive to the extraction of DNA, RNA, proteins and even metabolites. LM has been widely used by animal biologists (Isenberg et al. Citation1976) to study gene expression in specific cell types (Emmert-Buck et al. Citation1996) and to elucidate the associated molecular events (reviewed in Fink and Bohle Citation2005; Espina et al. Citation2006, Citation2007). In more recent years, LM has been adapted for plant tissues (Day et al. Citation2005, Citation2006; Nelson et al. Citation2006; Ramsay et al. Citation2006; Balestrini and Bonfante Citation2008). The advantage that this technology offers, with respect to other techniques that isolate homogeneous populations of a specific cell-type (i.e. cell sorting), is that LM can usually be applied to all cells that can be identified by conventional microscopy without the obligate use of specific cell markers or genetic lines (Birnbaum et al. Citation2003; Galbraith and Birnbaum Citation2006). The relative disadvantages of LM are of both a technical and technological nature. For example, histological fixation and processing are generally required and these can compromise the target materials (i.e. RNA, DNA, proteins, metabolites). A second problem is that the number of cells that can be recovered is limited by the abundance and recognition of the cellular targets in the histological sections (Nelson et al. Citation2006). Another limitation is that LM instrumentation is costly.
The aim of this short review is to summarize the applications of LM to the study of the interaction between a plant and both pathogenic and symbiotic organisms, giving also some information about the technical aspects concerning plant sample preparation.
How Laser Microdissection operates
The LM apparatus is generally attached to a light microscope, and tissue dissection is computer-controlled. Several instruments that isolate individual cells or groups of cells from intact tissues are commercially available. These microscopes are based on two methods of isolation: Laser Capture Microdissection (LCM) and laser cutting (Day et al. Citation2005; Nelson et al. Citation2006). In LCM, the considered cells are attached to a thermoplastic cap covered film using a pulsed infrared laser, which is manipulated so that it melts and fuses the film onto these cells. When the cap is removed, the target region is selectively pulled away from the surrounding tissues (Emmert-Buck et al. Citation1996). At the end of a session, the cap, containing the selected cells, is used for downstream applications (i.e. DNA, RNA extraction and molecular analyses). This was the first type of commercially available LM microscope and was produced by Arcturus (now Molecular Devices Corporation; Union City, USA, CA [http://www.moleculardevices.com/home.html]). An alternative microdissection approach, laser cutting, uses a UV laser to excise target regions from tissue sections. In these systems, a cursor is used to outline an area for microdissection. They have been designed in different ways in order to capture the microdissected cells. In one case, after microdissection, the excised fragment is catapulted upwards into a tube cap. This is called Laser Pressure Catapulting (LPC). This design was pioneered by P.A.L.M. Microlaser Technologies AG (Carl Zeiss MicroImaging; Munich, Germany [http://www.palm-microlaser.com/]). By contrast, in another microscope design, the sample simply falls by gravity into the collection tube without any extra forces. This system is called Laser Microdissection (LMD). This design is typical of microscopes such as those made by Leica (Leica Microsystems Inc., Germany [http://www.leica-microsystems.com]). The LPC and LMD systems allow the collection of a single cell as well as groups of cells or tissue regions. A new generation of LM systems includes both an infrared laser (as in an LCM) and a UV laser, thus allowing both laser-excised microdissection and capture. This latter type (Veritas Microdissection System) was designed by Molecular Devices Corporation (previously Arcturus). Specific cells can also be identified because the microscopes have been fitted with fluorescence optics. Although not essential, this could be very useful for marker or reporter-assisted identification of target cells (Tang et al. Citation2006). However, all LM methods of target isolation have been used successfully on plant materials.
Preparation of plant samples for LM
For LM, the tissues are first fixed and sectioned, and then the target cells are isolated from the non-target cells under a microscope. The fixation and embedding steps probably represent the most limiting factors to reach the final aim of good quality samples for molecular work. LM sample preparation requires a balance between two contrasting aims: to preserve enough visual detail to identify specific cells during harvesting, and to allow the maximum subsequent recovery of the material (i.e. nucleic acids, proteins and/or metabolites) from the harvested cells. The preparation of plant samples has been clearly illustrated in several original papers (Asano et al. Citation2002; Kerk et al. Citation2003; Klink et al. Citation2005; Inada and Wildermurth 2005; Tang et al. Citation2006; Cai and Lashbrook Citation2006) and in several reviews (Day et al. Citation2005, Citation2006; Nelson et al. Citation2006; Balestrini and Bonfante Citation2008; Gomez and Harrison Citation2009). Two methods have been utilized to prepare sample sections for LM: cryosectioning and paraffin sectioning. Cryosectioning is commonly used in animal research due to its speed and potentiality to better preserve intact molecules, including RNAs and proteins. In addition to the fact that the sample preparation for the sectioning is fast, the sample remains cold at all times and, presumably, this would decrease the probability of proteolysis or RNA degradation. However, it is not an easy task to obtain a plant tissue conservation that allows the identification of the selected cells. Freezing procedures can cause the formation of ice crystals inside the vacuoles and air spaces between cells in mature plant tissues: both these features compromise tissue histology, eventually leading to the disassembly of cell structure. The preparation of frozen sections of more mature or vacuolated plant material generally requires fixation as well as a cryoprotectant treatment, such as sucrose, in order to alleviate the tissue damage caused by freezing. The application of this method has so far been limited to prepare a few plant tissues for LM (Asano et al. Citation2002; Nakazono et al. Citation2003; Casson et al. Citation2005; Woll et al. Citation2005). As an alternative, where a more satisfactory preservation of tissue histology is required for target identification (i.e. in studies on plant interactions) samples are embedded in paraffin after fixation. Although this protocol provides excellent morphology, the RNA and protein yield is reduced compared with that from frozen samples: tissue fixation and paraffin embedding could in fact result in a considerable loss in quality and quantity of the extracted RNA. Nevertheless, satisfactory amounts of RNA have been obtained from paraffin-embedded material (Kerk et al. Citation2003; Tang et al. Citation2006) and the improved morphology is sometimes essential to identify the appropriate cell types. In addition, from paraffin embedded samples, it is possible to make perfect serial sections.
A key aspect of tissue preparation that affects the balance between morphological preservation and genomic/proteomic recovery is fixation. Two major types of fixatives have been used to prepare tissues for LM. Even if RNA isolated from archival formaldehyde-fixed animal tissues is routinely used in medical molecular diagnostic studies, particularly in cancer biology (Dillon et al. Citation2001), cross-linking fixatives (i.e. formalin) have been demonstrated to provide poor quality RNA from microdissected plant cells (Kerk et al. Citation2003). On the other side, precipitative fixatives, such as Farmer's fixative (ethanol/glacial acetic acid 3:1), are simple to make and provide both sample histological detail of plant tissues and enough RNA recovery for the downstream molecular analyses. RNA extracted from microdissected cells collected from plant tissues fixed with this method has been successfully used for molecular analyses (Kerk et al. Citation2003; Nakazono et al. Citation2003; Ramsay et al. Citation2004; Klink et al. Citation2005, Citation2007a; Ithal et al. Citation2007). A fixative that leads to a good balance between tissue preservation versus RNA recovery is a solution known as methacarn, which consists of absolute methanol-chloroform-glacial acetic acid (6:3:1; Takagi et al. Citation2004; Jiang et al. Citation2006; Balestrini et al. Citation2007; Guether et al. Citation2009). As an alternative, Inada and Wildermuth (Citation2005) introduced the use of a rapid microwave paraffin preparation method that avoids any fixative for the preparation of Arabidopsis leaf tissue for LM. This method led to sections with excellent preservation of internal leaf structures, while the extracted RNA was of sufficient yield to be used in downstream applications. As a further improvement, a method based on microwave-accelerated acetone fixation and paraffin-embedding has proved to preserve both tissue histology and protein fluorescence, leading to an RNA yield of suitable quality for microarray applications (Tang et al. Citation2006).
An important thing to remember about the application of LM is that no optimal protocol for all plant tissues exists. Therefore, the researcher should plan on optimizing the fixation and processing protocols on a case-by-case basis (Nelson et al. Citation2006). However, a number of protocols are now available providing a good starting point for most applications.
Application of LM in plant biology: Gene expression studies and proteomics
Although some applications of LM in combination with DNA analyses (Scutt et al. Citation1997; Meimberg et al. Citation2003; Hobza and Vyskot Citation2007) have been performed, most LM methods in plant researches have been developed for the identification of differentially expressed transcripts and are thus focused on LM-derived RNA, as reported in recent reviews published over the past few years (Day et al. Citation2005, Citation2006; Nelson et al. Citation2006; Ramsay et al. Citation2006; Ohtsu et al. Citation2007, Balestrini and Bonfante Citation2008). An important aspect of most LM-mediated gene expression research is that the RNA usually has to be amplified in order to obtain a sufficient amount of sample for the desired work. RNA yield obtained from microdissected cells is in fact generally enough for targeted molecular analysis like RT-PCR, while one or two rounds of RNA amplification are required to obtain the microgram amounts of mRNA needed for global profiling analyses (Nakazono et al. Citation2003; Klink et al. Citation2007a; Ithal et al. Citation2007). Several methods and commercial kits for RNA amplification, that maintain the relative representation of RNA species, have been developed and can be used specifically for this purpose (Brandt Citation2005). The most common method is based on in vitro transcription (IVT) (Petalidis et al. Citation2003). This linear amplification involves producing double-stranded cDNA with a T7 priming sequence at the 3' end. This template is then used by T7 RNA polymerase to generate copies of the cDNA template (Day et al. Citation2007). PCR-based procedures for RNA amplification have also been used, enabling much greater yields per round of amplification (Iscove et al. Citation2002). The PCR-based method in fact produces exponentially amplified cDNA template that then can be used for downstream analyses and, for this reason, less starting material can be used to generate enough target for high throughput analyses using only one round of amplification. PCR- and IVT-based amplification methods have recently been evaluated by Day et al. (Citation2007) in microarray experiments on laser microdissected endosperm. Starting from only 50 ng of RNA, amplification methods produced sufficient product for microarray hybridizations, with two-round IVT giving the best results and allowing the identification of endosperm enriched marker genes (Day et al. Citation2007). A T7 RNA polymerase-based RNA amplification was recently used to generate sufficient cDNA for 454 sequencing from RNA of maize SAM (Shoot Apical Meristem) cells isolated by LM (Emrich et al. Citation2007).
Proteins are the primary effectors of biological function in living organisms. It is therefore desirable to extend high throughput gene expression analyses to the protein level, especially since it has been demonstrated that RNA and protein levels do not always correlate (Liu et al. Citation2006; Dembinsky et al. Citation2007). However, only a few LM studies focused on protein (Schad et al. Citation2005a; Dembinsky et al. Citation2007) and metabolites have been so far carried out on plant materials (Schad et al. Citation2005b; Schneider and Hölscher Citation2006). Unlike transcript profiling –which can be performed from very small sample amounts due to efficient amplification strategies – no amplification procedure is possible for proteins. LM plant tissue-homogeneous samples have been used for protein identification either through two-dimensional gel electrophoresis (2-DE), or high-efficiency liquid chromatography (LC) in conjunction with tandem mass spectrometry (MS/MS) (Schad et al. Citation2005a). Dembinsky et al. (Citation2007) focused on examining protein in pericycle cells of maize, providing a comparative analysis between microarray outcomes and protein profiling. In detail, among the most abundant soluble pericycle proteins separated via two-dimensional electrophoresis, 20 proteins were identified via ESI MS/MS mass spectrometry, thus defining a first reference dataset of the maize pericycle proteome (Dembinsky et al. Citation2007). Metabolite profiling in a tissue- or cell-specific manner has been reported in combination with LC-MS/MS (Schad et al. Citation2005b) and cryogenic NMR spectrometry (Schneider and Hölscher Citation2006). In these studies, the preparation of sections is a crucial step as, when standard tissue fixation and embedding protocols are used to prepare histological sections, molecules, in particular metabolites, can be either efficiently extracted by dehydrating solvents or washed out by embedding agents. The possibility to use LM in combination with a proteomics/metabolomics approach would be an important advancement in plant-pathogen and plant-symbiotic interaction investigations.
Applications of LM to plant interaction studies
The study of plant-pathogen and plant-microbe interactions is surely an area where LM could have an impact since these experiments provide information about the spatial expression of the genes involved in interaction processes. The responses in these interactions can in fact be localized in specific cell types or microbial structures. So far only a few studies have been focused on the application of LM in plant-microbe interactions (Ramsay et al. Citation2004; Klink et al. Citation2005, Citation2007a; Tang et al. Citation2006; Balestrini et al. Citation2007) and these studies have focused on gene expression. When LM is used, cells associated with particular infection stages can be visualized under the microscope and harvested to verify the response of the plant in different cell types and during the progression of the infection. In the last few years, large amounts of data have been generated from microarray analyses of interactions between host plants and pathogenic (Klink et al. Citation2007a; Wise et al. Citation2007) or symbiotic organisms (Küster et al. Citation2007). However, most gene expression profiling studies rely on bulked material, and the analysis is carried out on samples from entire plants or whole organs. As a result, the data represents an average of many cell types, and gene expression data cannot be assigned to particular cell types (Tao et al. Citation2003). It is clear how, in the case of transcript and protein profiling, the LM approach could overcome the shortcomings of methods that use material isolated from whole organs.
Molecular studies of a biological event at single cell resolution provide more accurate information on genetic variations, development, and responses to biotic/abiotic stresses (Klink et al. Citation2005; Ramsay et al. Citation2006). Furthermore, bulk tissue analyses can mask important and unique localized responses. Several transcript profiling and other RNA analyses are currently underway that are investigating the interaction between plant and microbe through isolating the specific cells involved in the process. An important caveat is that it is essential to develop protocols and tools for LM-assisted analyses in order to examine specific plant cell populations through the progression of infection. This allows researchers to carry out spatial and temporal gene expression profiling (Day et al. Citation2005). These sorts of projects include investigations of the gene expression that occur during the interaction between plants and nematodes (Ramsay et al. Citation2004; Klink et al. Citation2005, Citation2007a; Ithal et al. Citation2007), fungal pathogen (Tang et al. Citation2006) and arbuscular mycorrhizal (AM) fungi (Balestrini et al. Citation2007; Guether et al. Citation2009; Gomez et al. Citation2009) (). Each of these studies, which so far have been performed in the field of plant/microbe interactions, employs an embedding step followed by serial sectioning of the tissue since the cells are located deep within the root tissue. These steps allow LM to have access to the target cells.
Table 1 List of papers on the application of LM in plant-nematode and plant-microbe interactions
Plant-pathogen interactions
Plant-nematode interactions
The infection of plants by parasitic nematodes is a major agricultural problem. Worldwide, this plant-pathogen interaction results in ~137 billion US dollars in lost revenue, annually (Abad et al. Citation2008). Thus, understanding the nature of disease progression is urgently needed. Establishment and development of infections by parasitic nematodes are complicated processes that involve an intimate interaction between the host and its pathogen. An area of interest is the development and maintenance of nematode feeding sites. LM methods that faithfully dissect out the feeding sites are important improvements to the study of this significant plant-pathogen interaction (Ramsay et al. Citation2004; Klink et al. Citation2005, Citation2007a; Ithal et al. Citation2007; Klink and Matthews Citation2008).
LM was used to collect cells specifically from earlier stages of infection in interactions between host plants and parasitic nematodes. For example, RNA was extracted 72–96 h after infection from giant cells induced in tomato roots by root-knot nematodes (Ramsay et al. Citation2004). These cells correspond, in fact, to provascular cells that are induced to re-enter the cell cycle. In those studies, the RNA was converted to cDNA. The analysis demonstrated amplified PCR products using primers designed for cell-cycle genes (Ramsay et al. Citation2004). A comparative analysis of giant cell gene expression was made to cDNA constructed from leaf material (Ramsay et al. Citation2004). They showed that differences existed in the quantity of PCR product on agarose gels between the giant cell and leaf tissue samples (Ramsay et al. Citation2004). The analyses did not provide a quantitative analysis of gene expression.
Infection of soybean (Glycine max) by the soybean cyst nematode (Heterodera glycines) is slightly different from the infection of roots by the root-knot nematode. During infection, the H. glycines penetrate the root and migrate toward the vascular tissue. They usually select a pericycle cell and then initiate the formation of a feeding cell by injecting substances into the selected cell. The nematode induces a variety of changes in the cell it selects as its feeding site, which results in the formation of a multinucleate feeding site (i.e. the syncytium). Thus, the syncytium is very different from the root cells it originates. Challenges to studying H. glycines infection at cellular resolution are that the infection process is not synchronous. The syncytial cell is also encased deeply within the root. It is clear, however, from previous anatomical observations (Endo Citation1965, Citation1991; Riggs et al. Citation1973; Kim et al. Citation1987), that gene expression within the syncytium is different from its neighboring cells and the cells from which it originated. The recalcitrance of their isolation forced many investigators to rely on whole roots as a source of RNA for gene expression studies (Khan et al. Citation2004; Klink et al. Citation2005, Citation2007a; Alkharouf et al. Citation2006). Physical separation and isolation of syncytia can, however, be performed by using LM (), and this provides a means of obtaining homogeneous cell samples and additional insights into their biology. In a first paper to report the use of LCM in examining syncytium gene expression, root samples enriched in syncytial cells were collected using LCM (Klink et al. Citation2005). RNA was extracted and used to make a cDNA library. Expressed sequence tags (ESTs) were produced and used for a Gene Ontology (GO) analysis. Many genes were identified, providing a new and more in-depth look at syncytial cells (Klink et al. Citation2005). Increases in transcript levels of some of these genes were confirmed by qRT-PCR and this was the first quantitative analysis of genes isolated by LM syncytia. Among them, an aquaporin (GmPIP2,2), α-tubulin (GmTubA1) and β-tubulin (GmTubB4). However, several other genes of unknown identity were also analyzed (Klink et al. Citation2005). Results were also confirming by using in situ hybridization and immunohistological techniques. In addition, the obtained cDNA has been used as a template to clone full-length cDNAs. Therefore, in those studies Klink et al. (Citation2005) established the protocols and procedures needed for gene expression experiments in syncytial cell samples.
Figure 1. (Color online). Microdissection of the syncytium formed by H glycines in the roots of G. max. (A) Eight day post infection (dpi) time point syncytial cell (white arrow) prior to microdissection identified by their proximity to H glycines (black arrowhead)s; (B) A syncytial cell after microdissection; microdissected syncytial cell (area between white arrows); nematode (black arrowhead) Bar, 100 µm

Recently, the same group has used LM to isolate homogeneous syncytium samples experiencing either a resistant or susceptible reaction (Klink et al. Citation2007a). The work was facilitated by the development of a commercial microarray (Affymetrix®) that could be used to examine G. max gene expression. In detail, the soybean GeneChip®, that contains 37,744 G. max probe sets representing 35,611 genes was used to study gene expression occurring inside the syncitial cell. In addition, an advantage in the analyses is that a single G. max genotype (i.e. Peking) could be used to obtain both resistant and susceptible reactions. The analysis provided numerous and intriguing insights into the biology of the syncytium, identifying gene expression that was unique to the syncytium. The authors tackled this problem through a series of computational analyses. Importantly, gene expression unique to the syncytium has been identified comparing syncytia to both whole root samples and LM-isolated pericycle cells. Many differentially expressed genes that were specific to the incompatible or compatible response within syncytium samples were identified. Among those highlighted in the incompatible response at three dpi were those encoding lipoxygenase, heat shock protein 70 and superoxide dismutase (Klink et al. Citation2007a). In addition, these experiments led to the identification of differentially expressed genes in the target cell that were not differentially expressed in a microarray analysis that examined gene expression in whole infected roots (Klink et al. Citation2007a, Citationb). To give an example of the number of genes differentially expressed, comparing gene expression of the syncytium undergoing a resistant reaction at three days post infection (dpi) to gene expression in whole root samples at the same time point, the analysis identified 147 genes that were induced only syncytium samples and 2360 genes that were induced only in whole root samples undergoing a resistant reaction. Only 19 genes were found to be induced and common between the syncytium and whole root samples. Suppressed genes were also identified: 278 genes that were suppressed only in syncytium samples undergoing a resistant reaction and 3624 genes that were suppressed only in 3 dpi whole root samples undergoing a resistant reaction. Only 39 genes were found to be suppressed and common between the syncytium and whole root samples undergoing a resistant reaction. The same analysis was performed also comparing gene expression of the syncytium undergoing a susceptible reaction, at 3 dpi and 8 dpi, respectively, to gene expression in whole root samples at the same time points. A complete description of the differentially expressed genes, at the different considered time points, can be found in Klink et al. (Citation2007a). An analysis that focused on gene expression during a susceptible response using the same system was also performed from another group (Ithal et al. Citation2007). In this last, an LCM system has been used instead of an LMD system as in Klink et al. (Citation2007a). The analysis of gene expression changes in developing syncytia, ranging from 0 to 10 days post-inoculation (dpi), led to the identification of 1,765 transcripts enriched or suppressed in syncytia at an early developmental time point compared with cells from a corresponding region of non-infected root tissues. Transcript abundance changes reflected alterations in cellular processes related to metabolism, defense, cell signaling, transport, and cell wall architecture (Ithal et al. Citation2007).
Taken together, these experiments have provided new insights into the complex molecular mechanisms underlying syncytium formation, and demonstrated that the mass of transcriptional activity in the whole root masks the identification of transcriptional events that occur within syncytial cells. Therefore, the use of LCM helped to de-obfuscate gene expression occurring in the syncytium from that which occurs in the surrounding cell types (i.e. cortex, root stele), thus showing that it is a useful tool to study plant-pathogen interactions.
Plant-pathogenic fungi interactions
In 2006, Tang et al. reported the first successful application of LM to the global gene expression analysis of early fungal-plant interactions. In that analysis, LM was applied to isolate individual maize cells associated with Colletotrichum graminicola hyphae at an early stage of infection. The LM-derived RNA was used to generate a global expression profile using a fungal microarray. Interestingly, an isolate of the anthracnose stalk rot fungus Colletotrichum graminicola expressing the Anemonia majano cyan fluorescent protein, was used to facilitate visual localization of fungal hyphae in planta. Compared to hand dissection or whole-tissue sampling, LM allows a much more homogeneous sampling of fungal developmental stages than gross tissue sampling, and is only limited by the ability to characterize and locate hyphal growth stages in fixed sections (Tang et al. Citation2006). A fixation method that could preserve tissue histology, protein fluorescence and also yielded RNA of a suitable quality for microarray applications was reported for the first time. The LM-derived RNA, even after two-rounds of linear amplification, was of sufficient quality and quantity for the identification of global expression profiling using a fungal microarray. The analysis identified 437 induced and 370 suppressed C. graminicola genes, respectively (Tang et al. Citation2006). The differential expression of several representative transcripts was confirmed by qRT-PCR. On the whole, the results demonstrated that the LM approach can reveal clues about the fungal genes that are involved in pathogenesis.
Another plant-pathogen interaction research field in which LM could be applied is the analyses of specific plant cell populations that are associated with the different infection stages of infection by biotrofic fungi (i.e. those belonging to the ascomycetes casual agent of powdery mildew that exclusively infects epidermal cells [Erysiphe spp.]). During infection, Erysiphe hyphae grow on the surface of the leaf. The hyphal tip then penetrates the epidermal cells, and originates a structure, called a haustorium, from which the fungus drags nutrients. The problem with studying this biological system is that the plant response occurs in both the infected epidermal cells, and in the neighboring uninfected mesophyll cells. Thus, LM could be of value and a powerful tool to isolate the several cell types involved in the interaction (Inada and Wildermurth 2005).
Plant-symbiotic interactions
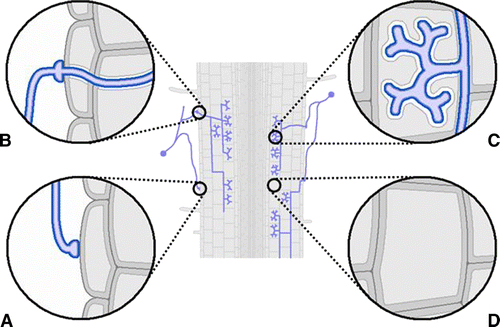
Arbuscular mycorrhizal (AM) fungi are an essential feature of the biology and ecology of most terrestrial plants. In AM symbiosis, the fungus obtains carbon from the plant, but, in the meantime, it transfers mineral nutrients from the soil to the root cells. During intracellular colonization, dramatic changes take place in the root structure. Recent observations have shown that the hypha enters through a single root epidermal cell, leading to the creation of a novel tunnel-like structure called the pre-penetration apparatus, PPA (Genre et al. Citation2005). This process is accompanied by important changes in gene expression (Siciliano et al. Citation2007). Subsequently, the cortical tissue is colonized with coils and intercellular hyphae which spread the infection. The fungus forms highly branched intracellular structures, called arbuscules, within the inner cortical cells. Arbuscules, which give the name to this symbiosis, are key structures of the AM symbiosis since they are considered the preferential site of the nutrient exchange between the host and symbiont. The fungus supplies the host with important mineral nutrients (i.e. phosphate [Pi]), while in return it receives carbon compounds. The accommodation of fungal hyphae inside root cortical cells leads to substantial changes in the cellular architecture of the root cortex cell. In addition to modifications shown by the nucleus, plastids, vacuoles and cytoskeleton, a new apoplastic compartment (i.e. the interface) is produced (Balestrini and Bonfante Citation2005). All of the cellular responses during this process are mirrored by important changes in gene expression. These gene expression changes have been revealed through global transcriptome analysis in Oryza (rice), Medicago and Lotus (Balestrini and Lanfranco Citation2006 [and references therein]; Guether et al. Citation2009). Therefore, global transcriptome profiling reveals the genes activated in mycorrhizal roots (Küster et al. Citation2007). However, these types of analyses suffer from the same problems mentioned in the previous sections. Transcript dilution occurs, making the identification of transcripts that pertain to the system difficult to identify. Therefore, whole root expression profiling is complicated by the presence of the multiple cell types that are involved in the interaction. For example, in a fully differentiated mycorrhizal root, various cell types are present (). These cells include colonized epidermal and cortical cells and, among them, non-colonized cells. There are also cells that have been colonized at different times during the spread of the infection. It is likely that each cell-type has a different role(s) in the interaction. Thus, examination of the process at the molecular level suffers from the drawback that the development of the symbiosis is not a synchronous process. After the initial colonization of the root cortex, secondary infection events commence and the colonization is reiterated.
Figure 2. (Color online). Scheme of a mycorrhizal root. Several cell types can be present: (A) Epidermal cell in contact with the fungal appressorium; (B) epidermal cell with an intracellular fungal hypha; (C) arbuscule-containing cells; (D) cortical non-colonized cell
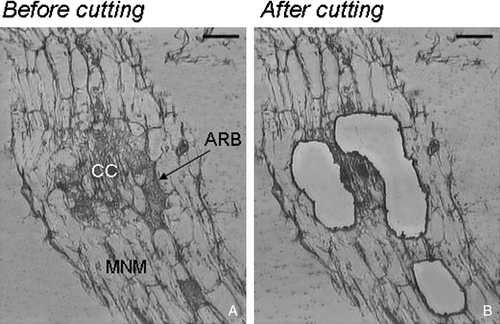
Over the last few years, LM has been used to study cell-specificity in arbuscular mycorrhizae. Particular attention has been paid to the cortical cells containing the arbuscules. In the first work published on the application of LM to AM interaction, a protocol for harvesting specific cell populations from paraffin sections of tomato mycorrhizal roots by the AS LMD system was developed by Balestrini et al. (Citation2007). The system was used to obtain cell samples that would yield RNA of sufficient quantity and quality for molecular analyses. The LM approach led to novel insights into the distribution of phosphate transporter (PT) transcripts during the AM interaction between tomato and Glomus mosseae. Transcripts of five tomato PT genes (LePTs) were, in fact, simultaneously detected in arbuscule-containing cells, unlike the neighboring non-colonized cells. On the fungal side, the H+ATPase (GmHA5) and the PT (GmosPT) mRNAs were exclusively found in arbusculated cells. These results suggest that plants ensure phosphate uptake through the functional redundancy of a gene family. Due to the crucial role of phosphorus (P) in their nutrition, plants seem to guarantee this function through the functional redundancy of a gene family. The contemporaneous presence of five PT mRNAs in the arbuscule-containing cells strongly suggests that the symbiosis enhances plant Pi uptake capabilities by recruiting additional PTs in this cell population. The discovery that five plant and one fungal PT genes are consistently expressed inside the arbusculated cells provides a new scenario for the plant-fungus nutrient exchanges (Balestrini et al. Citation2007). Recently, the gene expression of Lotus japonicus arbuscule-containing cells isolated by LM was validated to confirm array experiments (Guether et al. Citation2009). As in the previous work by Balestrini et al. (Citation2007), three types of homogeneous cell populations were microdissected. These cells included: (i) cortical cells from non-mycorrizal roots (C); (ii) non-colonized cortical cells from mycorrhizal roots (MNM); and (iii) arbuscule-containing cells (ARB) (). The RT-PCR reactions highlighted the presence of transcripts corresponding to seven genes with a role exclusively in arbuscule-containing cells (LjPT4 [phosphate transporter], LjCesA [cellulose synthase], LjMLO2 [MLO], LjCel1 [β-1,4-endoglucanase], LjMyb [Myb transcription factor like protein], LjPP [Protease inhibitor/seed storage/LipidTransfer Protein] and LjPTR [peptide transporter]). Taken together, these results provide novel information on the location of the genes that are potentially involved in the construction of interfacial compartment and/or in cell wall modification during intracellular colonization as well as in nutrient transport. The exclusive localization of LjPT4 in arbusculated cells was consistent with that reported for orthologs in Medicago (Harrison et al. Citation2002) and tomato (Balestrini et al. Citation2007).
Figure 3. Example of Laser Microdissection of arbusculated cortical cells: Lotus japonicus mycorrhizal roots. (A) Before laser; (B) tissue remaining after cutting Bars, 25 µm
Our observations also suggest that, in rare cases, the response in MNM cell-type populations can vary among the different biological replicates. This result could be due to the so far undescribed heterogeneity of such a cell population and agrees with the observations of Genre et al. (Citation2008), who recently reported how some cortical cells respond to the presence of the fungus before arbuscule development, while other cells are not activated. The slight variability observed in the molecular analyses is probably due to the fact that the MNM cell type population could be formed by both the cortical cells that have already been activated and also by those that have not.
However, the epidermis, due to its location at the interface between the soil and the inner tissues of the root, also plays a crucial role in both mineral nutrition and signalling events. Therefore, the gene expression profile of epidermal cells could provide a new tool to help understand the role played by this cell layer in AM associations. It is well known that root hairs, which are specialized epidermal cells, are key structures in mineral uptake and, at the same time, epidermal cells are crucial in mediating signals from the extraradical fungus (Parniske Citation2008). The finding of epidermal marker genes could be important to study cell-specificity associated to epidermal cells in order to have a control for the collected material. RT-PCR experiments with specific primers for a tomato extensin gene (LeEXT1) and an RNaseLX gene (Köck et al. Citation2006) have shown that LM allows transcripts to be localized in epidermal cells and that, according to Bucher et al. (Citation2002), LeEXT1 can be considered an epidermal marker in tomato (RB and JGA, unpublished results).
These promising results demonstrate that LM is a suitable technology to investigate whether epidermal and cortical cells, but also other cell-types (i.e. central cylinder), accomplish different functions during the building up of a functioning symbiotic root. A further step will be to obtain ‘global’ profiles associated with the different cell-types that are present in mycorrhizal roots (i.e. arbusculated cells).
Conclusions and perspectives
LM provides significant advantages for researchers studying plant-pathogenic and plant-symbiotic interactions. With the isolation of specific target cells, host plant responses can be investigated much more accurately in terms of regulation and/or compartmentalization of gene expression. This technology, in combination with large-scale genomic analyses (i.e. microarray hybridizations, cDNA library construction, GS FLX454 technology), will be very useful to identify genetic determinants involved in several processes, including signalling, nutrient transfer, transcription, cell wall modifications and defence that occur during these interactions (Ohtsu et al. Citation2007). LM has already been used successfully, in combination with microarray experiments, to obtain gene expression profiles in several plants during different developing stages and in plant-nematode interactions undergoing susceptible and resistant reactions (Klink et al. Citation2007a).
Recently, the coupling of LCM and GS FLX454 (454 Life Sciences; Branford, CT) sequencing technologies (LCM-454) has been demonstrated in studies on shoot apical meristem cells of maize (Emrich et al. Citation2007). The work resulted in the identification of > 261,000 ESTs. It was found that > 68,000 of the > 261,000 sequences (∼26.05%) identified in that analysis did not match any of the > 648,000 ESTs that were previously deposited in the public databases (Emrich et al. Citation2007). Thus, the combination of LM and 454 sequencing resulted in the identification of numerous rare and possibly cell-type-specific RNAs that were unidentified, previously (Emrich et al. Citation2007). This approach could be very useful for AM interactions, as well as for other plant interactions, and provide information on gene expression in both partners.
The application of a protocol for paraffin embedding, such as that proposed by Tang et al. (Citation2006), which maintains eGFP fluorescence for the visualization of cells and sufficient RNA quality for the molecular analysis, could be very useful in the early stages of the interaction. The use of the eGFP technology would allow the collection of only the specific cells that respond before or at the moment of contact.
The LM application has also been proposed for other interactions (i.e. plant–plant pathogenic interactions (Pérez-de-Luque et al. Citation2008) and root nodules). Considering the tissue and cellular complexity of an ectomycorrhiza, LM technology could also be very useful to collect the two specific fungal compartments separately (the mantle and the Hartig net) which are expected to be functionally different, at least in terms of nutrient uptake and transport (Nehls et al. Citation2001).
The application of LM to the study of plant biology and plant interactions has so far been devoted mainly to cell-specific gene expression profiling. However, protocols regarding LM in combination with protein/metabolite analyses will surely be developed.
Acknowledgements
The contributions to this review have in part been funded by the Regione Piemonte Tech4wine Project to RB; Integral Project and the Italian-Spanish project Azioni Integrate to PB; IPP-CNR. JGA has been supported by a Lagrange fellowship of the Fondazione CRT. VPK thankfully acknowledges the United Soybean Board for their continued support and a Research Initiation Program Grant from Mississippi State University.
References
- Abad , P , Gouzy , J , Aury , JM , Castagnone-Sereno , P , Danchin , EG , Deleury , E , Perfus-Barbeoch , L , Anthouard , V , Artiguenave , F , Blok , VC , Caillaud , MC , Coutinho , PM , Dasilva , C , De Luca , F , Deau , F , Esquibet , M , Flutre , T , Goldstone , JV , Hamamouch , N , Hewezi , T , Jaillon , O , Jubin , C , Leonetti , P , Magliano , M , Maier , TR , Markov , GV , McVeigh , P , Pesole , G , Poulain , J , Robinson-Rechavi , M , Sallet , E , Ségurens , B , Steinbach , D , Tytgat , T , Ugarte , E , van Ghelder , C , Veronico , P , Baum , TJ , Blaxter , M , Bleve-Zacheo , T , Davis , EL , Ewbank , JJ , Favery , B , Grenier , E , Henrissat , B , Jones , JT , Laudet , V , Maule , AG , Quesneville , H , Rosso , MN , Schiex , T , Smant , G , Weissenbach , J and Wincker , P . 2008 . Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita . Nat Biotech , 26 : 909 – 915 .
- Alkharouf , NW , Klink , VP , Chouikha , IB , Beard , HS , MacDonald , MH , Meyer , S , Knap , HT , Khan , R and Matthews , BF . 2006 . Timecourse microarray analyses reveals global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode) . Planta , 224 : 838 – 852 .
- Asano , T , Masumura , T , Kusano , H , Kikuchi , S , Kurita , A , Shimada , H and Kadowaki , K . 2002 . Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: Toward comprehensive analysis of the genes expressed in the rice phloem . Plant J , 32 : 401 – 408 .
- Balestrini , R and Bonfante , P . 2005 . The interface compartment in arbuscular mycorrhizae: A special type of plant cell wall? . Plant Biosyst , 139 : 8 – 15 .
- Balestrini , R and Lanfranco , L . 2006 . Fungal and plant gene expression in arbuscular mycorrhizal symbiosis . Mycorrhiza , 16 : 509 – 524 .
- Balestrini , R , Gómez-Ariza , J , Lanfranco , L and Bonfante , P . 2007 . Laser Microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells . Mol Plant-Microbe Interact , 20 ( 9 ) : 1055 – 1062 .
- Balestrini , R and Bonfante , P . 2008 . Laser Microdissection (LM): Applications to plant materials . Plant Biosyst , 142 : 331 – 336 .
- Birnbaum , K , Shasha , DE , Wang , JY , Jung , JW , Lambert , GM , Galbraith , DW and Benfey , PN . 2003 . A gene expression map of the Arabidopsis root . Science , 302 : 1956 – 1960 .
- Brandt , SP . 2005 . Microgenomics: gene expression analysis at the tissue-specific and single-cell levels . J Exp Bot , 56 : 495 – 505 .
- Bucher , M , Brunner , S , Zimmermann , P , Zardi , GI , Amrhein , N , Willmitzer , L and Riesmeier , JW . 2002 . The expression of an extensin-like protein correlates with cellular tip growth in tomato1 . Plant Physiol , 128 : 911 – 923 .
- Cai , S and Lashbrook , CC . 2006 . Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling . Plant J , 48 : 628 – 637 .
- Casson , S , Spencer , M , Walker , K and Lindsey , K . 2005 . Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis . Plant J , 42 : 111 – 123 .
- Day , RC , Grossniklaus , U and Macknight , RC . 2005 . Be more specific! Laser-assisted microdissection of plant cells . Trends Plant Sci , 10 : 397 – 405 .
- Day , RC , McNoe , LA and Macknight , RC . 2006 . Transcript analysis of laser microdissected plant cells . Physiol Plant , 129 ( 2 ) : 267 – 282 .
- Day RC McNoe L Macknight RC 2007 Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser-microdissected endosperm Int J Plant Genomics
- Dembinsky , D , Woll , C , Saleem , M , Liu , Y , Fu , Y , Borsuk , LA , Lamkemeyer , T , Fladerer , C , Madlung , J , Barbazuk , B , Nordheim , A , Nettleton , D , Schnable , PS and Hochholdinger , F . 2007 . Transcriptomic and proteomic analyses of pericycle cells of the maize (Zea mays L) primary root . Plant Physiol , 145 : 575 – 588 .
- Dillon , D , Zheng , K and Costa , J . 2001 . Rapid, efficient genotyping of clinical tumor samples by laser-capture microdissection/PCR/SSCP . Exp Mol Pathol , 70 : 195 – 200 .
- Emmert-Buck , MR , Bonner , RF , Smith , PD , Chuaqui , RF , Zhuang , Z , Goldstein , SR , Weiss , RA and Liotta , LA . 1996 . Laser capture microdissection . Science , 274 : 998 – 1001 .
- Emrich , SJ , Barbazuk , WB , Li , L and Schnable , PS . 2007 . Gene discovery and annotation using LCM-454 transcriptome sequencing . Genome Res. , 17 : 69 – 73 .
- Endo , BY . 1965 . Histological responses of resistant and susceptible soybean varieties, and backcross progeny to entry development of Heterodera glycines . Phytopathology , 55 : 375 – 381 .
- Endo , BY . 1991 . Ultrastructure of initial responses of resistant and susceptible soybean roots to infection by Heterodera glycines . Rev Nematol , 14 : 73 – 94 .
- Espina , V , Milia , J , Wu , G , Cowherd , S and Liotta , LA . 2006 . Laser capture microdissection . Methods Mol Biol , 319 : 213 – 229 .
- Espina , V , Heiby , M , Pierobon , M and Liotta , LA . 2007 . Laser capture microdissection technology . Expert Rev Mol Diagn , 7 : 647 – 657 .
- Fink , L and Bohle , RM . 2005 . Laser microdissection and RNA analysis . Methods Mol Biol , 293 : 167 – 185 .
- Galbraith , DW and Birnbaum , K . 2006 . Global studies of cell type-specific gene expression in plants . Ann Rev Plant Biol , 57 : 451 – 475 .
- Genre , A , Chabaud , M , Timmers , T , Bonfante , P and Barker , DG . 2005 . Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection . Plant Cell , 17 : 3489 – 99 .
- Genre , A , Chabaud , M , Faccio , A , Barker , DG and Bonfante , P . 2008 . Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota . Plant Cell , 20 : 1407 – 1420 .
- Gomez SK , Javot H , Deewatthanawong P , Torres-Jerez I , Tang Y , Blancaflor EB , Udvardi MK , Harrison MJ . 2009 . Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis . BMC Plant Biol . 9 : 10 doi 10.1186/1471-2229-10 .
- Gomez SK , Harrison MJ . 2009 . Laser microdissection and its application to analyze gene expression in arbuscular mycorrhizal symbiosis . Pest Manag Sci . www.interscience.wiley.com. doi 10.1002/ps.1715
- Guether M , Balestrini R , Hannah M , He J , Udvardi M , Bonfante P 2009 Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus New Phytol doi: 101111/j1469-8137200802725x In press
- Harrison , MJ , Dewbre , GR and Liu , J . 2002 . A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi . Plant Cell , 14 : 2413 – 2429 .
- Haseloff , J , Siemering , KR , Prasher , DC and Hodge , S . 1997 . Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly . PNAS , 94 : 2122 – 2127 .
- Hobza , R and Vyskot , B . 2007 . Laser microdissection-based analysis of plant sex chromosomes . Methods Cell Biol , 82 : 433 – 453 .
- Inada , N and Wildemuth , MC . 2005 . Novel tissue preparation method and cell-specific marker for laser microdissection of Arabidopsis mature leaf . Planta , 221 : 9 – 16 .
- Iscove , NN , Barbara , M , Gu , M , Gibson , M , Modi , C and Winegarden , N . 2002 . Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA . Nat Biotech , 20 : 940 – 943 .
- Isenberg , G , Bielser , W , Meier-Ruge , W and Remy , E . 1976 . Cell surgery by laser micro-dissection: A preparative method . J Microsc , 107 : 19 – 24 .
- Ithal , N , Recknor , J , Nettleton , D , Maier , T , Baum , TJ and Mitchum , MG . 2007 . Developmental transcript profiling of cyst nematode feeding cells in soybean roots . Mol Plant-Microbe Interac , 20 : 510 – 525 .
- Jefferson , RA , Kavanagh , TA and Bevan , MW . 1987 . GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants . EMBO J , 6 : 3901 – 3907 .
- Jiang , K , Zhang , S , Lee , S , Tsai , G , Kim , K , Huang , H , Chilcott , C , Zhu , T and Feldman , LJ . 2006 . Transcription profile analyses identify genes and pathways central to root cap functions in maize . Plant Mol Biol , 60 : 343 – 363 .
- Kerk , NM , Ceserani , T , Tausta , SL , Sussex , IM and Nelson , TM . 2003 . Laser capture microdissection of cells from plant tissues . Plant Physiol , 132 : 27 – 35 .
- Kim , YH , Riggs , RD and Kim , KS . 1987 . Structural changes associated with resistance of soybean to Heterodera glycines . J Nematol , 19 : 177 – 187 .
- Khan , R , Alkharouf , N , Beard , HS , MacDonald , M , Chouikha , C , Meyer , S , Grefenstette , J , Knap , H and Matthews , B . 2004 . Microarray analysis of gene expression in soybean roots susceptible to the soybean cyst nematode two days post invasion . J Nematol , 36 : 241 – 248 .
- Klink , VP , Alkharouf , N , MacDonald , M and Matthews , B . 2005 . Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode) . Plant Mol Biol , 59 : 965 – 979 .
- Klink , VP , Overall , CC , Alkharouf , NW , MacDonald , MH and Matthews , BF . 2007a . Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines) . Planta , 226 : 1389 – 1409 .
- Klink , VP , Overall , CC , Alkharouf , NW , MacDonald , MH and Matthews , BF . 2007b . A time-course comparative microarray analysis of an incompatible and compatible response by Glycine max (soybean) to Heterodera glycines (soybean cyst nematode) infection . Planta , 226 : 1423 – 1447 .
- Klink , VP and Matthews , BF . 2008 . The use of laser capture microdissection to study the infection of Glycine max (soybean) by Heterodera glycines (soybean cyst nematode) . Plant Signaling Behav , 3 : 206
- Köck , M , Stenzel , I and Zimmer , A . 2006 . Tissue-specific expression of tomato Ribonuclease LX during phosphate starvation-induced root growth . J Exp Bot , 57 : 3717 – 3726 .
- Küster , H , Vieweg , MF , Manthey , K , Baier , MC , Hohnjec , N and Perlick , AM . 2007 . Identification and expression regulation of symbiotically activated legume genes . Phytochem , 68 : 8 – 18 .
- Liu , Y , Lamkemeyer , T , Jakob , A , Mi , G , Zhang , F , Nordheim , A and Hochholdinger , F . 2006 . Comparative proteome analyses of maize (Zea mays L) primary roots prior to lateral root initiation reveal differential protein expression in the lateral root initiation mutant rum1 . Proteomics , 6 : 4300 – 4308 .
- Meimberg , H , Thalhammer , S , Brachmann , A , Muller , B , Eichacker , LA , Heckl , WM and Heubl , G . 2003 . Selection of chloroplasts by laser microbeam microdissection for single-chloroplast PCR . Biotechniques , 34 : 1238 – 1243 .
- Nakazono , M , Qiu , F , Borsuk , LA and Schable , PS . 2003 . Laser capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: Identification of genes expressed differentially in epidermal cells or vascular tissue of maize . Plant Cell , 15 : 583 – 596 .
- Nehls , U , Mikolajewski , S , Magel , E and Hampp , R . 2001 . Carbohydrate metabolism in ectomycorrhizas: gene expression, monosaccharide transport and metabolic control . New Phytol , 150 : 533 – 541 .
- Nelson , T , Tausta , SL , Gandotra , N and Liu , T . 2006 . Laser microdissection of plant tissue: What you see is what you get . Ann Rev Plant Biol , 57 : 181 – 201 .
- Ohtsu , K , Takahashi , H , Schnable , PS and Nakazono , M . 2007 . Cell type-specific gene expression profiling in plants by using a combination of laser microdissection and high-throughput technologies . Plant Cell Physiol , 48 : 3 – 7 .
- Parniske , M . 2008 . Arbuscular mycorrhiza: the mother of plant root endosymbioses . Nat Rev Microbiol , 6 : 763 – 775 .
- Pérez-de-Luque , A , Moreno , MT and Rubiales , D. 2008 . Host plant resistance against broomrapes (Orobanche spp): Defence reactions and mechanisms of resistance . Ann Appl Biol , 152 : 131 – 141 .
- Petalidis , L , Bhattacharyya , S , Morris , GA , Collins , VP , Freeman , TC and Lyons , PA . 2003 . Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis . Nucleic Acids Res , 31 ( 22 ) : 142
- Ramsay , K , Wang , Z and Jones , MGK . 2004 . Using laser capture microdissection to study gene expression in early stages of giant cells induced by root-knot nematodes . Mol Plant Pathol , 5 : 587 – 592 .
- Ramsay , K , Jones , MGK and Wang , Z . 2006 . Laser capture microdissection: A novel approach to microanalysis of plant-microbe interactions . Mol Plant Pathol , 7 : 429 – 43 .
- Riggs , RD , Kim , KS and Gipson , I . 1973 . Ultrastructural changes in Peking soybeans infected with Heterodera glycines . Phytopathol , 63 : 76 – 84 .
- Schad , M , Lipton , MS , Giavalisco , P , Smith , RD and Kehr , J . 2005a . Evaluation of two-dimensional electrophoresis and liquid chromatography–tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples . Electrophoresis , 26 : 2729 – 2738 .
- Schad , M , Mungur , R , Fiehn , O and Kehr , J . 2005b . Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana . Plant Meth , 1 : 2
- Schneider , B and Hölscher , D . 2006 . Laser microdissection and cryogenic nuclear magnetic resonance spectroscopy: An alliance for cell type-specific metabolite profiling . Planta , 225 : 763 – 770 .
- Scutt , CP , Kamisugi , Y , Sakai , F and Gilmartin , PM . 1997 . Laser isolation of plant sex chromosomes: Studies on the DNA composition of the X and Y sex chromosomes of Silene latifolia . Genome , 40 : 705 – 715 .
- Siciliano , V , Genre , A , Balestrini , R , Cappellazzo , G , DeWitt , P and Bonfante , P . 2007 . Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus . Plant Physiol , 144 : 1455 – 1466 .
- Takagi , H , Shibutani , M , Kato , N , Fujita , H , Lee , K-Y , Takigami , S , Mitsumori , K and Hirose , M . 2004 . Microdissected region-specific gene expression analysis with methacarn-fixed, paraffin-embedded tissues by real-time RT-PCR . J Histochem Cytochem , 52 : 903 – 913 .
- Tang , W , Coughlan , S , Crane , E , Beatty , M and Duvick , J . 2006 . The application of laser microdissection to in planta gene expression profiling of the maize anthracnose stalk rot fungus Colletotrichum graminicola . Mol Plant-Microbe Interact , 19 : 1240 – 1250 .
- Tao , Y , Xie , Z , Chen , W , Glazebrook , J , Chang , HS , Han , B , Zhu , T , Zou , G and Katagiri , F . 2003 . Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell , 15 : 317 – 330 .
- Wise , RP , Moscou , MJ , Bogdanove , AJ and Whitham , SA . 2007 . Transcripts profiling in host-pathogen interactions . Annu Rev Phytopathol , 45 : 329 – 369 .
- Woll , K , Borsuk , LA , Stransky , H , Nettleton , D , Schnable , PS and Hochholdinger , F . 2005 . Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1 . Plant Physiol , 139 : 1255 – 1267 .