ABSTRACT
Since the Green Revolution, intensive application of agrochemicals has increased productivity in agriculture, at a great cost in terms of water pollution, loss of soil fertility and biodiversity, and negative effects on human health. Scientific advance and increasing public awareness are driving a change toward sustainable practices. In such a context, the symbiosis between plants and arbuscular mycorrhizal (AM) fungi is extremely promising: AM interaction improves plant mineral nutrition and stress tolerance. In turn, AM fungi receive plant photosynthesis-derived carbon. A complex chemical dialogue mediates plant-fungus recognition and symbiosis establishment: AM fungi perceive root-secreted strigolactones, which promote spore germination, hyphal growth, branching and metabolism. Host roots recognize their symbionts through chitin-derived molecules. Such Myc–factors activate a range of symbiotic responses, preparing the plant to a successful association. Here we review the most recent advances in knowledge of AM signaling molecules, with a focus on their possible application.
Introduction
Over the last decades agricultural practices involved a misuse of pesticides and chemical fertilizers, aiming to combat pests and to increase productivity to feed the growing human population. Such phytochemical treatments are causing a range of diverse drawbacks, including water pollution, loss of soil fertility and biodiversity, as well as food poisoning, leading to several detrimental effects on human health (Vassilev et al. Citation2015). A shift toward low-input, environmentally-friendly practices to promote biodiversity and sustainability is urgent and more and more led by public awareness. In this scenario, beneficial plant-microbe interactions are raising increasing interest. Among them, arbuscular mycorrhizal (AM) symbiosis is one of the most widespread (around 72% of plant species), moreover, it involves the majority of crop species (Gutjahr and Parniske Citation2013). AM symbiosis improves plant mineral absorption (namely phosphorus and nitrogen), tolerance to biotic and abiotic stresses and overall fitness, while fungi are rewarded with carbon compounds derived from the photosynthetic process, such as sugars and lipids (Rich et al. Citation2017). During the early stages of interaction, AM fungi recognize host plants through the perception of root-secreted strigolactones (SLs; Akiyama et al. Citation2005; Boyno and Demir Citation2022). SLs boost spore germination, hyphal growth and branching (Akiyama et al. Citation2005; Besserer et al. Citation2006), mitochondrial activity in the presymbiotic mycelium (Besserer et al. Citation2006), expression of effector genes (Tsuzuki et al. Citation2016), as well as the release of plant-directed chemical signals (Genre et al. Citation2013), known as Myc-factors (Boyno and Demir Citation2022). Currently characterized Myc-factors are chitin-derived molecules: chito-oligosaccharides (COs; Genre et al. Citation2013) and lipo-chito-oligosaccharides (LCOs; Maillet et al. Citation2011). Myc-factor perception activates plant symbiotic responses, including a massive cell structural reorganization, in order to favor the subsequent fungal accomodation (Gutjahr and Parniske Citation2013). Recent works have demonstrated that exogenous CO application in Oryza sativa and Medicago truncatula induced AM symbiosis signaling, stimulated lateral root branching and AM symbiosis development (Oldroyd Citation2013; Feng et al. Citation2019; Volpe et al. Citation2020; Crosino et al. Citation2021), leading the way to the applicative use of such treatments in sustainable agriculture (Lanfranco et al. Citation2016).
Here we will discuss the latest advances in our understanding of the nature, role and applicative potential of signaling molecules involved in AM establishment.
Strigolactones
Strigolactones (SLs) are a small group of molecules released by roots of different plants, stimulating seed germination in weeds of Striga (also known as witchweeds), a genus composed by hemiparasite plants (Al-Babili and Bouwmeester Citation2015). The first natural SLs were isolated from cotton root exudates (Gossypium hirsutum), strigol and strigyl acetate, able to stimulate seed germination in the hemi-parasitic plant Striga lutea (Cook et al. Citation1966). Since then, over 20 SLs have been characterized in root exudates of several plants, including maize (Zea mays), red clover (Trifolium pratense) and sorghum (Sorghum bicolor) (Hauck et al. Citation1992; Müller et al. Citation1992; Siame et al. Citation1993; Yokota et al. Citation1998; Xie et al. Citation2013). These molecules are known to induce germination in different genera of parasitic weeds besides Striga, such as Orobanche, Alectra, and Phelipanche (Yoneyama et al. Citation2013). The estimated damage caused by parasitic weeds for rice crops in Africa alone is around 200 million USD, increasing by 30 million each year (Rodenburg Citation2016), and over 60 million hectares of farmland worldwide are infested by Striga and Orobanche species, causing a loss quantified in billions of dollars every year (Parker Citation2009). Such discoveries led to a fascinating question on the reason why host plants produce compounds which stimulate their own parasitization. The answer to this question came in 2005 when the benefits of SL exudation for plants were clarified. In fact, an extensive hyphal branching in germinating spores of the AM fungus Gigaspora margarita was highlighted upon treatment with the natural strigolactones 5-deoxy-strigol, sorgolactone, strigol and GR24, a synthetic analog (Akiyama et al. Citation2005). Only later, SL role as plant hormones was outlined, initially highlighting their function as shoot branching inhibitors (Gomez-Roldan et al. Citation2008; Umehara et al. Citation2008). Subsequently, a broader range of hormonal effects was discovered, including regulation of root architecture, secondary growth promotion, leaf senescence regulation and coordination of root responses during nutrient deficiency (Yoneyama et al. Citation2007; Umehara et al. Citation2010; Kohlen et al. Citation2011; Niu et al. Citation2013).
Structure
Strigolactones (SLs) are a group of molecules composed of a tricyclic lactone (ABC) linked by an enol-ether bond to a butenolide moiety (). Many natural and synthetic SL compounds present different variation in the ABC structures, such as avenaol, debranones, 5-deoxystrigol and carlactone. From such differences, two main classes of plant SLs were generated, based on stereochemical modifications in the B-C link: strigol and orobanchol type. Strigol-like SLs are characterized by the β orientation (up; 8bS configuration) of the C ring, whereas orobanchol-like SLs contain an alpha-oriented (down; 8bR configuration) C ring.
Figure 1. Classification and chemical structure of Strigolactones. These are generally composed of a tricyclic lactone (ABC) connected, by an enol-ether bond, to a butenolide moiety (red), which represents the most conserved trait of the molecule between plant taxa.
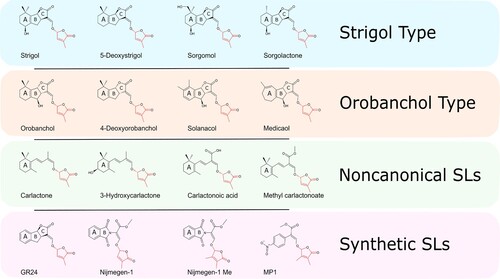
SLs that present the entire ABC-ring system are called canonical, in contrast to those lacking A, B, or C-rings, that are known as noncanonical. An example of such noncanonical SLs are avenaol, heliolactone and lotuslactone characterized from root exudates of wild oat, sunflower and Lotus japonicus respectively (Yoneyama et al. Citation2020).
The only part of the molecule that appears to be conserved across all bioactive SLs is the D ring with 2’R configuration. In fact, several studies have demonstrated that SL activity is lost when the enol-ether-connected D ring section is modified (Kapulnik et al. Citation2011; Boyer et al. Citation2012, Citation2014; Fukui et al. Citation2013; Rasmussen et al. Citation2013; Abe et al. Citation2014; Prandi et al. Citation2014; Ueno et al. Citation2014; Fridlender et al. Citation2015; Tsuchiya et al. Citation2015; De Saint Germain et al. Citation2016).
The majority of plant species contain a single type of molecule as the major SL component in root exudates. An exception is represented by tobacco, which exudes significant quantities of both strigol and orobanchol (Xie et al. Citation2013), making the investigation of SL biosynthesis and transport particularly interesting for this species.
Biosynthesis
Even if a few steps in SL biosynthesis remain unclear, our current knowledge was built using information from different plants, which partially differ in their metabolic pathways. As depicted in , SL biosynthesis starts from trans- β-carotene, which is converted to 9-cis-β-carotene by the isomerase DWARF27 (D27; Alder et al. Citation2012). Subsequently, CAROTENOID CLEAVAGE DIOXYGENASE7 (CCD7) and CCD8 complete the formation and orientation of the D ring, generating the inactive SL common precursor carlactone, which lacks the tricyclic structures and presents a β-ionone ring instead.
Figure 2. General scheme of Strigolactone biosynthesis. Strigolactones are synthesized from trans-β-carotene and the large diversity of SL structures is obtained thanks to the recruitment of different enzymes in different plant species.
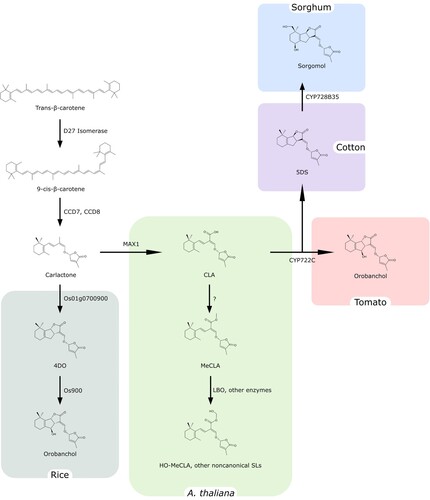
In Arabidopsis, MORE AXILLARY GROWTH1 (MAX1), from the CYP711A family of cytochromes P450, converts carlactone to carlactonoic acid (CLA), which is subsequently methylated by an unknown enzyme to produce MeCLA (Abe et al. Citation2014). Rice has five homologs of MAX1 with different functions. One of them, Os01g0700900, oxidizes carlactone to 4-deoxyorobanchol (4DO), the precursor for orobanchol-type SLs. The subsequent conversion from 4DO to orobanchol is carried out by Os900 and the other rice MAX1 homologs (Zhang et al. Citation2014).
In contast to rice, CLA is directly converted into orobanchol in tomato, which only has one MAX1 homolog and is unable to produce orobanchol from 4DO (Yoneyama et al. Citation2018). A transcriptomic approach in cowpea recently identified the enzyme responsible for such direct conversion from CLA to orobanchol, as CYP722C, which belongs to a distant cytochrome P450 clade from MAX1 (Wakabayashi et al. Citation2019). However, in cotton, CYP722C converts CLA to 5-deoxystrigol (5DS) (Wakabayashi et al. Citation2020), while CYP728B35 (from an additional P450 clade), produces sorgomol from 5DS in sorghum (Wakabayashi et al. Citation2021).
Besides the cytochromes P450 family, other enzymes have been related to the diversification of SL family members. Among them, LATERAL BRANCHING OXIDOREDUCTASE (LBO), a 2-oxoglutarate-dependent dioxygenase, is strongly co-expressed with canonical SL biosynthetic genes in transcriptomic and reverse genetic experiments in A. thaliana plants (Brewer et al. Citation2016). Furthermore, Brewer et al. (Citation2016) highlighted a possible conversion from MeCLA to hydroxymethyl carlactonoate in in vitro experiments with extracts from Escherichia coli expressing LBO. LBO-like proteins are present in the majority of land plants, and even if no lbo mutants have been characterized so far (Walker et al. Citation2019), the lack of lotuslactone production in L. japonicus mutant for a 2-oxoglutarate-dependent dioxygenase, related to LBO, validates the implication of this molecular family in noncanonical SL biosynthesis (Mori et al. Citation2020).
In short, the canonical SL production from CLA is regulated by three different P450 clades, while MeCLA appears to be essential for the synthesis of noncanonical SLs, such as heliolactone (Wakabayashi et al. Citation2020).
Redundancy in gene functions and parallel metabolic pathways - whereby mutants in individual biosynthetic genes display a phenotype, if any – limits the use of forward genetics to elucidate SL biosynthesis (Brewer et al. Citation2016). A major advance came from the introduction of transcriptomics and reverse genetics studies (Yoneyama and Brewer Citation2021), which provided a broader view of the biosynthesis of structurally unrelated SLs. The characterization of a great variety in SL composition within crop plant exudates, in combination with the knowledge derived from such new approaches, should increase the potential to manipulate SL diversity in crops, in order to boost agronomic performance in the near future (Chesterfield et al. Citation2020).
SL perception
In plants, the hormonal function of SL is dependent on their perception by DWARF14 (D14), an α/β hydrolase that binds canonical SLs and splits them into hydroxymethylbutenolide (HMB) and an ABC ring formyltricyclic lactone (ABC-FTL, ; Hamiaux et al. Citation2012; Zhao et al. Citation2013; Nakamura et al. Citation2013). On this event, D14 undergoes a conformational change, ascribable to either the unbroken SL or to one of its hydrolysis products (De Saint Germain et al. Citation2016; Yao et al. Citation2016; Seto et al. Citation2019; Lee et al. Citation2020). This promotes the association of D14, D3 (AtMAX2; an F-box protein, part of the Skp1-Cullin-F-box complex) and D53 (homolog of SUPPRESSOR OF MAX2 1 in A. thaliana) in a complex. As a consequence, D53 undergoes ubiquitination and is degraded, ending its activity of downstream signaling inhibition (Hamiaux et al. Citation2012; Zhou et al. Citation2013) and allowing D14 proteasomal degradation (Chevalier et al. Citation2014). In addition, D3 was also reported to bind D14, inhibiting its hydrolytic activity (Shabek et al. Citation2018).
Figure 3. Perception of Strigolactones and signaling. Strigolactones are perceived by D14, which forms a complex with D3 and D53, a negative regulator of downstream signaling. The molecule is then hydrolyzed to hydroxymethylbutenolide (HMB) and an ABC ring formyltricyclic lactone (ABC-FTL), while D14 and D53 are degraded, allowing the activation of downstream signaling.
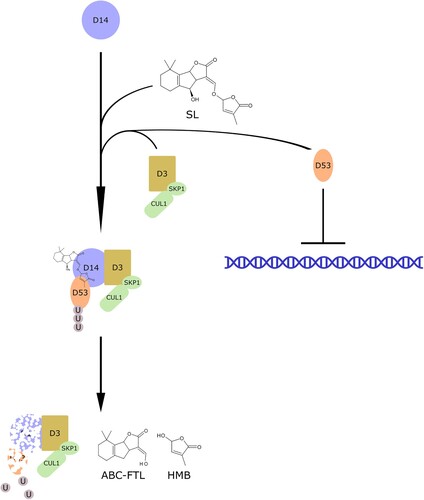
An important cross-talk between SLs and karrikins has been recently identified. Karrikins are compounds derived from cellulose combustion, which are implicated in the germination of dormant seeds after a bush-fire (Flematti et al. Citation2015). Such molecules are detected by a group of receptors called KAI2/HTL-type, that are involved in seed germination and seedling growth. It has been proposed that karrikins mimic KAI2 ligand (KL), a putative endogenous ligand involved in early plant development, sharing part of the signaling pathway with SLs (Conn and Nelson Citation2016). In fact, in parasitic plants, SL perception takes place via an HTL-type α/β hydrolase, a paralog of D14. HTL7 from S. hermonthica has the ability to hydrolyze SLs, generating an active intermediate essential for the transduction pathway that triggers seed germination (Shahul Hameed et al. Citation2018). However, due to their great variety in structure, only a fraction of SLs molecules activates the karrikin signaling pathway. Scaffidi et al. (Citation2014) highlighted a preferential affinity between synthetic SL isomers and KAI2 receptor, underlying a relationship between the recognition of SLs by D14 or KAI2 and the structural variations in the chiral carbon orientations at the junction of the BC and D rings. Downstream of D3, karrikin signaling pathway differentiates from SLs signaling for the degradation of SUPPRESSOR OF MAX2 1 (SMAX1) and SMAX1-LIKE2 (SMXL2) (Stanga et al. Citation2016).
SL perception in fungi remains largely unclear and has only been demonstrated in plant pathogenic fungi. In fact, mutant screening analyses highlighted the production of reactive oxygen species in response to SL treatment of Botrytis cinerea and Cryphonectria parasitica (Belmondo et al. Citation2017). Further investigations showed the up-regulation of genes involved in nuclear and cellular compartment reorganization, DNA-related functions and lipid metabolism after the application of GR24, a synthetic SL analog (Lanfranco et al. Citation2018). No evidence of D14-related genes has been found in the genomes of AM fungal symbionts and no alternative SL receptor has been characterized (Moscatiello et al. Citation2014; Chen et al. Citation2018). Nonetheless, regardless of their perception mechanism, SLs were shown to activate calcium-mediated signaling in the AM fungus G. margarita (Moscatiello et al. Citation2014).
In conclusion, SL perception mechanisms appear to be mediated by different receptors in each group of organisms. Thorough elucidation of such mechanisms represents a major milestone on the way to their application in sustainable agriculture as modulators of specific plant interactions with beneficial as well as pathogenic or parasitic organisms (Chesterfield et al. Citation2020).
Role of SL in AM symbiosis
Growing evidence is indicating that differences in SL bioactivity in each organism are largely related to their molecular structure. In line with that, a conserved AB-ring structure was suggested to be essential to trigger AM hyphal branching, one of the most striking and studied SL-dependent responses (Akiyama et al. Citation2005; Besserer et al. Citation2006). Furthermore, Akiyama et al. (Citation2010) demonstrated that an α-oriented hydroxyl group insertion at the C4 position of the B-ring boosted the hyphal branching effect, suggesting the importance of this functional group as hydrogen bond donor during SL interaction with the putative fungal receptor.
Additional SL-induced responses in AM fungi include the proliferation of nuclei in hyphal tips, the activation of lipid catabolism, a boost in mitochondrial activity and the release of chito-oligosaccharides (Besserer et al. Citation2006, Citation2008; Genre et al., Citation2013; Belmondo et al. Citation2017; Lanfranco et al. Citation2018). SLs were also reported to enhance hyphopodium formation on the host root surface (Kobae et al., Citation2018).
SL production and field application
In the light of such multifaced effects on plant health, different attempts were performed in the last few years to achieve large-scale SL production for field application. Firstly, SL application was intended as a strategy to limit the onset of parasitic plants, the so-called suicidal seed germination. This technique consists in SL field treatment before crop cultivation, with the purpose of inducing the germination of parasitic weed seeds that, in absence of a host, face the death. In sorghum, T-010, a synthetic SL, decreased the emergence of S. hermonthica by 94–100% in pot trials and by around 30% in field, with concomitant increase in sorghum head yield of almost 200% (Samejima et al., Citation2016). In the same way, Zwanenburg et al. (Citation2016), using the SL analogs Nijmegen-1 and Nijmegen-1 Me in tobacco crops, highlighted a reduction in O. ramosa emergence by at least 60% in six out of nine field trials. A further confirmation of the SL efficiency in suicidal seed germination comes from the work by Kountche et al. (Citation2019), where the authors developed a protocol for Striga control in rain-fed African fields. The use of Nijmegen-1, MP1 and MP3 in sorghum and pearl millet, resulted in a drastic reduction of Striga emergence by 55% and 65%, respectively. Such studies point out suicidal seed germination as a valid method to control parasitic weed infestations and a potent and critical instrument that, in the future, could be embraced in the areas characterized by the most severe Striga and Orobanche infestations, such as sub-Saharan Africa.
Following the discovery of SL role in AM establishment, SL application to promote AM development in agronomic contexts raised great interest. Nevertheless, even if SLs are of common use for research purposes and field trials, their introduction in agriculture and commercial release have not been achieved yet, largely due to difficulties in setting up their large scale production. Secondly, SL molecular instability, mainly due to the cleavage of the enol ether bond by nucleophilic agents such as water, makes commercial formulations, long-term storage and agronomic applications very difficult or ineffective (Aliche et al., Citation2020; Chesterfield et al., Citation2020). SL instability is a major issue under field conditions, such as soil pH and temperature (Zwanenburg & Pospíšil, Citation2013). The persistence of GR24 – which has better stability than natural compounds – is limited to 6–8 days in acidic soils and drops to an average of 1–3 days in alkaline soils (Miyakawa et al., Citation2019). Formulations that improve SL stability have been developed for Nijmegen-1 and its 3’-Me congener: the use of polyoxyethylene sorbitol hexaoleate as a stabilizer allowed field trials in tobacco plants infested by O. ramosa (Zwanenburg & Pospısil, Citation2013). In short, additional studies will be necessary to develop specific formulations for each synthetic SL, extending its stability and uptake and reducing soil leaching, while also preventing environmental pollution with the selected stabilizers and their own effects on AM development (Aliche et al., Citation2020). A third problem comes from SL-dependent stimulation of parasitic weed germination. A possible solution to this comes from our understanding of SL diversity. Sorghum mutants for LOW GERMINATION STIMULANT 1 (LGS1) are resistant to Striga hermonthica and Striga asiatica infection. This was related to a change in SL profile, with a reduction in strigol and an increase in orobanchol content (Gobena, et al., Citation2017). Interestingly, lgs1 mutants are not affected in their mycorrhization level compared to wild-type sorghum. Similarly, a Striga-resistant maize cultivar with a reduced level of 5-deoxystrigol and an increase in sorgomol exudation also had AM colonization levels comparable to wild-type plants (Yoneyama et al., Citation2015). Differently, a complete incapability to establish AM symbiosis was seen in the rice mutant d14-l (AtKAI2 homolog) instead, showing a total absence of hyphopodia formation (Gutjahr et al., Citation2015). On the contrary, the rice d14 mutant presented normal or even slightly higher level of AM colonization compared to WT plants (Yoshida et al., Citation2012). A middle ground was represented by mutants defective in downstream signaling components; rice d3 and pea rms4, with low levels of arbuscule and vesicle formation (Yoshida et al., Citation2012; Foo et al., Citation2013). Being D3/RMS4 a common protein for both SL and karrikin signaling pathways, it is easy to assume that the karrikin signaling activation is essential for AM symbiosis establishment. Following the indication of Scaffidi et al. (Citation2014), it could be interesting to evaluate if the preferential affinity of some SL compounds for KAI2 is reflected in higher AM colonization levels. All these observations, combined with the discoveries of Akiyama et al. (Citation2010), have raised awareness that changes in SL chemical structure have a deep impact on their effectiveness towards either symbiotic fungi or parasitic weeds, paving the way to the identification and design of specific SL treatments aimed at either promoting AM colonization or counteracting parasitic weed infection.
The very limited accumulation of SLs in plant tissues and exudates makes their extraction from plant materials not practical. For this reason, SLs are mainly produced by chemical synthesis. Unfortunately, the relative complexity of SL chemical structure requires over 20 steps, and also critically limits yield in the current production systems (Arshad & Frankenberger, Citation2002). A promising alternative is represented by the synthesis of just the bioactive part of the molecule. Such SL analogs can be synthesized in higher amounts and currently represent the most common source of SLs for research scopes (Chesterfield et al., Citation2020). An additional advantage of this method – whenever a clear structure-response relationship has been elucidated – is the possibility to exclusively produce the chemical structure that is responsible for the desired function (e.g. AM promotion, suicidal seed germination or drought stress tolerance, another effect of SLs). The most common SL analogs are currently Nijmegen-1 (Nefkens et al., Citation1997), GR24 (Besserer et al. Citation2008), CISA-1 (Rasmussen et al. Citation2013), sphynolacten-7 (Uraguchi et al. Citation2018) and strigolactams (De Mesmaeker et al. Citation2019), which are mainly used for suicidal seed germination. Notwithstanding such promising advantages, production costs remain a limiting factor for process scale-up. Furthermore, SL and SL analog chemical synthesis generates 1:1 racemic mixtures, in which only one of the enantiomers is biologically active (weakening the overall effectiveness), or each isomer presents distinct biological activities, generating unwanted effects. A well characterized example is GR24, which is 100 times less efficient than natural SLs in enhancing Orobanche minor seed germination (Yoneyama et al. Citation2010). On the same line, Akiyama et al. (Citation2010), found that (+)-5DS and (+)-GR24 were respectively 10 and 100-fold more effective in stimulating hyphal branching in AM fungi than their corresponding enantiomers.
Larger amounts of bioactive SLs might be obtained at smaller costs with the use of biosynthetic enzymes through microbial engineering. The central point of this approach is to reconstruct the SL biosynthetic pathway in a heterologous context, through the genetic transformation of bacterial or yeast strains. In addition, the use of enzymes catalyzing each biosynthetic reaction gives microbial engineering the potential to produce enantiomerically pure molecules, an outstanding advantage compared to chemical synthesis (Chesterfield et al. Citation2020). A promising example of this approach was recently presented by Wu (Citation2021), who successfully reconstituted CL, CLA, 4DO, 5DS and orobanchol biosynthesis in Escherichia coli – Saccharomyces cerevisiae consortia. Even if the yields of current attempts remain low, the optimization of microbial enzyme activity can be envisaged by gene editing. Lastly, this approach is also emerging as an accessible tool to functionally characterize SL biosynthetic enzymes, in analogy with the earlier development of enzyme-based industrial production of other valuable compounds, such as isoprenoids (Vickers et al. Citation2015).
Myc-factors
On the fungal side of the AM symbiotic dialogue, two structurally related classes of signaling molecules have been characterized in AM fungal exudates, stimulating symbiotic responses in their host plants: Myc-LCOs (Maillet et al. Citation2011) and Myc-COs (Genre et al. Citation2013). LCOs have been first characterized in rhizobium/legume symbiotic nitrogen fixation as bacterial signaling molecules (Nod-factors), whose recognition by the host plant is a prerequisite to symbiosis establishment (Denarie and Cullimore Citation1993). Studies on legumes have been fundamental to discover relevant similarities in molecular and cellular mechanisms between rhizobial and AM symbioses. The most striking example is represented by the common symbiotic signaling pathway (CSSP), a signaling pathway shared among such two mutualistic interactions, including the generation of repeated oscillations in nuclear Ca2+ concentration in plant cells. These similarities opened the way to the investigation of signal molecules produced by AM fungi, which led to the detection of sulphated and nonsulphated LCOs, structurally related to Nod-factors (Maillet et al. Citation2011), and short-chain chitin oligomers (COs; Genre et al. Citation2013).
Structure
COs are chitin/chitosan derivatives (), composed of N-acetylglucosamine (GlcNAc) and glucosamine (Glc) residues linked through a β(1→4) bond (Mourya et al. Citation2011; Lodhi et al. Citation2014). CO chemical properties and bioactivity are mostly influenced by their degree of polymerization (DP, number of GlcNAc/Glc residues), acetylation degree (AD, ratio between GlcNAc and total residues) and pattern of acetylation (PA, position of the acetylated residues within the molecule). Short-chain COs (with DP ≤ 8) are easily soluble in aqueous media (Hao Citation2021), and include the best characterized and most active Myc-factors, CO4 and CO5 (Genre et al. Citation2013).
Figure 4. Chemical structure of chitin-derived fungal signals. Myc-factors characterized in arbuscular mycorrhizal fungi include (A) tetra-chito-oligosaccharide, composed of four monomers of N-acetylglucosamine (GlcNAc), and (B) tetra-lipo-chito-oligosaccharide bearing a 16:1 fatty acid chain (red) and a carbamoyl group (blue). (C) octa-chito-oligosaccharide (CO8) are common elicitors of defense responses.
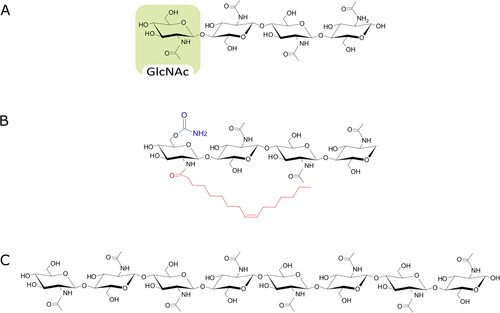
LCOs () are short-chain chitin oligomers substituted with a fatty acid and different chemical groups at the nonreducing end (Khokhani et al. Citation2021). The most common fatty acids in fungal LCOs are C16:0, C18:0, C18:1, in contrast with C16:1 and C20:2 acyl groups commonly found in Nod-factors. Most of the remaining substitutions (acetyl, methyl, sulphate and fucosyl sulphate groups) are common to both microbial signals, while the presence of an arabinose residue is unique to Nod-factors (Dénarié et al. Citation1996; Rush et al. Citation2020).
Biosynthesis
Rush et al. (Citation2020) found that 53 out of 59 fungal species tested, spread across most fungal phyla, were able to produce LCOs and the majority of fungal LCOs analyzed were similar to those exuded by AM fungi. Such work highlighted LCO production as a highly conserved trait in the fungal kingdom. However, very little is known about LCO biosynthesis in fungi.
In fact, most of our knowledge of the metabolic reactions leading to LCO biogenesis comes from studies in N-fixing rhizobia, where the activation of the chitin synthase NodC leads to the assembly of the chitin backbone. Following a few substitutions (such as fucosylation), the chitin deacetylase NodB removes the acetyl group from the nonreducing end of the molecule. This is followed by the action of NodA, which adds the acyl chain to the CO deacetylated site, before additional substitutions (e.g. methylation, carbamoylation, arabinosylation) take place (Poinsot et al. Citation2016). On this basis, LCO biosynthesis in fungi has been suggested to involve the action of different endochitinases, cleaving short-chain COs from longer cell wall-associated chitin chains, and acyltransferases binding the acyl chain (Khokhani et al. Citation2021).
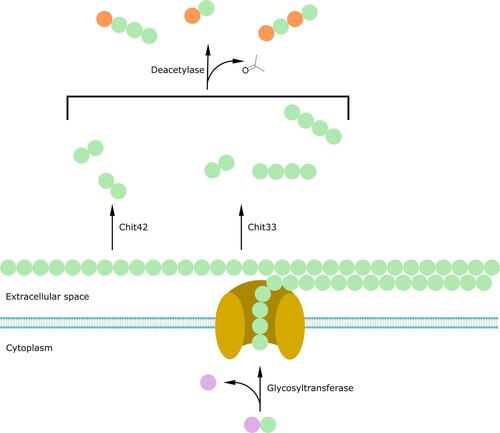
By contrast, CO biosynthesis in fungi has been elucidated more clearly (). Chitin is the main structural polymer of fungal cell wall. UDP-GlcNAc is used as substrate by cell membrane-associated chitin synthase complexes and added to the nonreducing end of the lengthening chitin chain. The resulting chitin chains are extruded in the extracellular space via a cell membrane channel composed of chitin synthase transmembrane domains. Lastly, extruded chitin chains combine into parallel and antiparallel fibrils through hydrogen bonds (Merzendorfer Citation2011; Orlean and Funai Citation2019). The subsequent synergic activity of chitinases and monoxygenases generates reducing and nonreducing ends in each chitin chain. These grant exochitinases access, whose activity causes CO cleavage (Langner and Göhre Citation2016). Different chitinases may generate COs of different DP. For example, in Trichoderma harzianum, both Chit33 and Chit42 produce CO2 while Chit33 can also produce CO4 (Berlemont Citation2017; Khokhani et al. Citation2021). Both chitin chains and COs can then be deacetylated by fungal chitin deacetylases, generating chitosan and COs with different AD (Grifoll-Romero et al. Citation2018).
Figure 5. Fungal biosynthesis of chito-oligosaccharides. N-acetylglucosamine (GlcNAc) monomers are transferred from UDP-GlcNAc to the forming chitin chain by the action of the glycosyltransferase and the chitin synthase (yellow). Chitin chains extruded in the extracellular space through a channel composed of chitin synthase transmembrane domains can then be cleaved by several chitinases (Chit33, Chit42) to produce chito-oligosaccharides of different length. Finally, a deacetylase can further remove acetyl groups from the newly formed oligomers. Purple = UDP; green = GlcNAc monomers; orange = Glc monomers.
Perception
In AM host plants, CO4/5 are perceived via plasma membrane-associated receptor-like kinase (RLK) complexes, which have been partially characterized in different species. In a recent study, He et al. (Citation2019) demonstrated the ability of the rice LysM-RLK OsLYK2 to specifically recognize CO4/5 (but not longer chain COs or LCOs) forming a complex with a second LysM-RLK, CHITIN ELICITOR RECEPTOR KINASE 1 (OsCERK1) and activate it by phosphorylation. A third member of the RLK complex, SYMBIOSIS RECEPTOR-LIKE KINASE (SYMRK/DMI2), much more specific for Myc-LCOs, was characterized in legumes. In M. truncatula, its activation upon LCO binding triggers the so-called common symbiosis signaling pathway, or CSSP, together with LYK3 and NFP (; Zipfel and Oldroyd Citation2017; Choi et al. Citation2018; Feng et al. Citation2019).
Figure 6. Myc-factor perception. (A) Studies in rice demonstrated that COs are perceived by OsLYK2 and OsCERK1, which is activated by phosphorylation. Investigations in M. truncatula showed that CO and LCO perception requires the action of DMI2, LYK3 and NFP. DMI2 binds to HMGR1, which starts mevalonate production and downstream signaling. This includes nuclear Ca2+ spiking, which also requires the activity of a few nuclear envelope-localized proteins: the cationic channel DMI1, the cyclic nucleotide-gated Ca2+ channel CNGC15s, the MCA8 Ca2+ pump and several nucleoporins in the nuclear pore complex. Ca2+ spiking is further decoded by the calcium and calmodulin-dependent kinase DMI3, which phosphorylates the transcriptional regulator MtIPD3, activating in turn DELLA, DIP1, NSP2, NSP1 and RAM1 transcription factors and driving the expression of AM-related genes. (B) Perception of alternative AM fungal signals by D14-L. After the perception of an unidentified ligand, D14-L form a complex with D3, driving the ubiquitination of the negative regulator SMAX1 and thus allowing downstream signaling culminating in gene regulation and symbiotic plant responses.
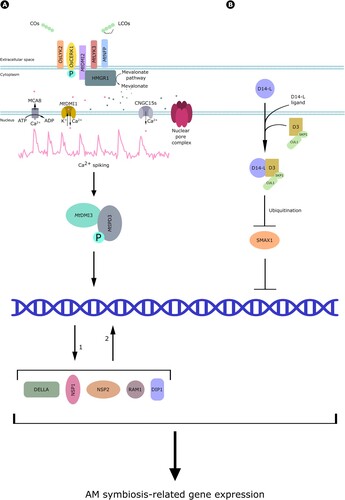
Little is known about the signal transduction from the plasma membrane to the nuclear envelope. One model suggests the association between SYMRK and the enzyme 3-hydroxy-3-methylglutaryl-CoA reductase 1 (HMGR1), essential for mevalonate synthesis (Kevei et al. Citation2007). Indeed, exogenous mevalonate application was shown to trigger downstream CSSP signaling such as nuclear Ca2+ spiking in the root epidermis (Venkateshwaran et al. Citation2015). In non-legumes, the complex formed between OsCERK1 and OsLYK2 seems to be essential to trigger Ca2+ spiking, since Oslyk2 mutants showed decreased levels of symbiotic responses (He et al. Citation2019). This induction of repeated oscillations in nuclear Ca2+ concentration is mediated by the action of a few nuclear envelope-localized proteins, including cationic channels such as MtDMI1, LjCASTOR, LjPOLLUX, the cyclic nucleotide-gated Ca2+ channel CNGC15s, and the MtMCA8 Ca2+ pump (Ané et al. Citation2002; Imaizumi-Anraku et al. Citation2005; Peiter et al. Citation2007; Venkateshwaran et al. Citation2012). The generation of Ca2+ spiking also requires NUCLEAR PORE COMPLEX PROTEIN 85 (NUP85), NUP133 and NENA, a WD40 repeat nucleoporin, most likely involved in the positioning of the above mentioned channels and pumps on the inner nuclear membrane (Kanamori et al. Citation2006; Saito et al. Citation2007; Groth et al., Citation2010).
Nuclear Ca2+ spiking is believed to be sensed by the calcium – and calmodulin-dependent serine/threonine protein kinase (CCaMK/MtDMI3), whose activation causes dimerization with the transcriptional activator MtIPD3/LjCYCLOPS. In turn, CYCLOPS phosphorylation boosts the activation of several transcription factors, including DELLA, DELLA Interacting Protein 1 (DIP1), Nodulation Signaling Pathway 2 (NSP2), NSP1 and Reduced Arbuscular Mycorrhiza 1 (RAM1), which drive the subsequent expression of AM symbiosis-related genes (Yu et al. Citation2014; Jin et al. Citation2016; Pimprikar et al. Citation2016; Zipfel and Oldroyd Citation2017).
On the edge between symbiosis and defense
As highlighted in the previous sections, COs are perceived by plants through the CSSP, activating symbiotic outcomes. However, since COs are chitin derivatives and chitin is the main structural polymer of the majority of fungi, COs are produced by a great variety of fungi, including plant pathogens (Rush et al. Citation2020). Plant developed the ability to activate immune responses through the recognition of specific non-self-molecules, called PAMPs (pathogen-associated molecular pattern). Concerning fungal pathogens, part of such PAMPs are represented by fungal cell wall-derived COs, resulting from the action of plant or fungal chitinases during plant tissue infection (Kasprzewska Citation2003; Lee et al. Citation2008; Wan et al. Citation2008; Volk et al. Citation2019). The most supported hypothesis explaining how chitin-derived molecules may trigger either symbiotic or defense responses considers CO length: COs with DP≥6 (typically CO8) are more prone to activate the plant immune response, whereas shorter COs are associated to symbiotic signaling (Shimizu et al. Citation2010; Nasir et al. Citation2018; Feng et al. Citation2019). This ability of root cells to discriminate CO length is believed to depend on the composition of LysM-RLK complexes. In a recent model by Chiu and Paszkowski (Citation2021) the ability of OsCERK1 to participate both in defense and in symbiotic responses is highlighted, and a mechanism of action, also involving OsMYR1 is proposed. In this model, the OsCERK1-OsMYR1 complex forms in the presence of CO4/5 or LCOs, leading to the subsequent activation of symbiotic responses. By contrast, the presence of CO8 triggers OsCERK1-OsCEBiP (CHITIN ELICITOR-BINDING PROTEIN) complex formation, eliciting chitin-triggered immunity and driving the production of reactive oxygen species and the phosphorylation of mitogen-activated protein kinases (MAPKs). In a recent review, Yang et al. (Citation2022) proposed an updated model, where two OsCEBiP monomers dimerize after binding to the same CO7-8 molecule, causing the recruitment and homodimerization of OsCERK1. Besides OsCEBiP, two additional effectors can bind long COs, OsLYP4 and OsLYP6, which are required to boost immunity outcomes (Yang et al. Citation2022). However, this model cannot be extended to all plants. In tomato, the OsCERK1 homolog SlLYK12 is almost entirely associated to symbiotic signaling, while the activation of the defense responses is dependent on SlLYK1 (Liao et al. Citation2018), indicating that the CERK1 homologs faced different duplication and subfunctionalization events in different families and species.
In spite of a broad set of experimental support, this model contrasts with a few recent observations. Feng et al. (Citation2019) reported CSSP activation upon CO8 application. They also suggested that a CO/LCO mix can be required for a fully efficient symbiotic outcome. Furthermore, Khokhani et al. (Citation2021) suggested three additional signaling mechanisms that may contribute to the successful stimulation of plant symbiotic responses. Firstly, high phosphate availability can activate plant immunity in the presence of any plant-fungus interaction and cause the degradation of DELLA transcription factors, thus inhibiting AM development. Secondly, the mechanical stimuli caused on the root epidermis by AM fungal hyphopodia or pathogenic fungal appressoria may also have a deep impact on downstream signaling. Lastly, the combination of chitin-derived signals with yet-to-identify molecules exclusive of AM fungi could also contribute to symbiont recognition. This latter hypothesis appears to be in line with the increasing evidence in favor of CSSP-independent signaling in AM symbiosis.
CSSP independent signaling
Besides the discrimination between symbiotic and pathogenic signals, a further level of complexity is present in legumes, where the distinction between structurally-similar LCOs of different origins (Nod-LCOs vs. Myc-LCOs) has to activate specific responses. This raised the hypothesis that additional signaling pathways exist, acting in parallel with the CSSP, as proposed by Bonfante and Requena (Citation2011). On this line, Gutjahr et al. (Citation2015) demonstrated the involvement of D14-L/KAI2 in Myc-factor perception. In this study, rice d14-l mutants were completely unable to establish AM interaction; furthermore, a drastically impaired AM colonization was also observed in mutants in D3, the protein that acts immediately downstream of D14-L (Yoshida et al. Citation2012).
A study conducted in rice by Choi et al. (Citation2020) described the transcriptional repressor SUPPRESSOR OF MAX2 1 (OsSMAX1) as an additional component of this signaling pathway, downstream of D3. When inoculated with R. irregularis, Ossmax1 mutants exhibited a higher level of colonization, overexpression of SL biosynthesis and AM-related genes, as well as increase in SL production, compared to WT plants. In addition, a restoration of WT mycorrhization level was achieved in d14-l/smax1 double mutants. The resulting model suggests that, upon the perception of a yet-unknown fungal signal, D14-L forms a complex with D3 which suppresses SMAX1 and positively regulates AM-related genes. Such studies have only started to shed light on this novel D14-L pathway and additional studies are required to identify upstream signals (and their fungal or endogenous origin), downstream molecular actors and the possible cross-talk with the CSSP, for example by analyzing Ca2+ spiking activation in different mutants.
Myc-factor production and applications
Irrespectively of the characterization of additional AM fungal signals and a number of studies showing that the perception of raw fungal exudates stimulates symbiotic responses in the host root (Gutjahr et al. Citation2008; Mukherjee and Ané Citation2011), the effect of currently known Myc-factors on AM development are being elucidated and the exogenous application of either COs or LCOs was shown to promote AM-related gene regulation and root colonization in different host plants. Sulfated LCOs were suggested to be more effective in stimulating symbiotic gene regulation in M. truncatula, compared to non-sulphated LCOs and COs; a combination of COs and LCOs was most effective in Lotus japonicus, while only CO4 produced a significant effect in rice (Sun et al. Citation2015; Feng et al. Citation2019). Lastly, a recent study showed that the application of purified short-chain COs to M. truncatula caused a global promotion of root colonization by the AM fungus (Volpe et al. Citation2020).
These results encouraged the development of Myc-factor production and application for the promotion of AM symbiosis in sustainable agricultural practices. Since Myc-factor isolation from AM fungi is not suitable for their large-scale production, three alternative strategies have been pursued.
Chemical approaches
Chemical synthesis of Myc-factors appears as a very versatile approach, allowing the synthesis of COs of any length and AD, not to mention the possibility to add the lateral substitutions typical of LCOs. Nevertheless, an alternation of protection and deprotection steps is required to prevent the formation of unwanted glycosidic bonds between monomers (Naqvi and Moerschbacher Citation2015; Hao Citation2021). Furthermore, the use of GlcNAc donors may cause the formation of abundant side products, such as 1,2-O, N-oxazoline (Hao Citation2021). Overall, even if significant improvements have been introduced over the last few years, including new iterative glycosylation methods and automated synthesis, several remaining disadvantages, such as product contamination and low yield, still limit the interest in synthetic approaches to large scale and low-cost Myc-factor production (Yamago et al. Citation2004).
As an alternative to chemical synthesis, chitin depolymerization by acid hydrolysis (Domard and Carter Citation1989; Kazami et al. Citation2015; Crosino et al. Citation2021), nitrous acid deamination (Tømmeraas et al. Citation2001), fluorolysis (Bosso et al. Citation1986; Defaye et al. Citation1994) or oxidative-reductive reactions with hydrogen peroxide (Tian et al. Citation2003) represent potentially easier and less expensive approaches to Myc-factor production. Large amounts of chitin can be obtained from waste products of fishing and food industry (crustacean exoskeletons and fungal biomass derived from the production of edible mushrooms are rich in chitin). A general drawback of these techniques is the alteration of the chitin chain reducing ends, which has a negative impact on the bioactivity of short chain COs. Furthermore, acid hydrolysis can modify CO acetylation degree (Naqvi and Moerschbacher Citation2015), which also affects their activity as symbiotic signals (Crosino et al. Citation2021). Lastly, the use of strong acids and their disposal in wastes raises major environmental concerns. To overcome such problems and improve chitin fibril exposure in the starting materials, a few authors experimented with diverse physical pre – or co-treatments, including ultrasonication (Takahashi et al. Citation1995; Machová et al. Citation1999; Ajavakom et al. Citation2012; Zhang et al. Citation2021), microwave irradiation (Roy et al. Citation2003; Wu et al. Citation2011; Ajavakom et al. Citation2012), supercritical water and mechano-chemical grinding (Savage Citation1999; Osada et al. Citation2013), gamma irradiation (Kang et al. Citation2007) and steam explosion (Alvira et al. Citation2016). Other studies have tested relatively less toxic solvents, such as ionic liquids or molten salts (Wilkes Citation2004; Passos et al. Citation2014; Vekariya Citation2017). At present, CO production from chitin is an achievable strategy, but its economic and ecological costs remain a major limitation.
Enzymatic approaches
A more environment-friendly approach to CO production is by using isolated enzymes. Indeed, both CO synthesis and chitin depolymerization can be achieved via enzymatic methods. GlcNAc residues and short chain COs can be assembled through the transglycosylation (TG) activity of chitinolytic enzymes, such as chitinases and lysozymes (Ling et al. Citation2018). β-NAcetylhexosaminidase from Aspergillus oryzae has been successfully employed to synthesize fully-acetylated COs (Singh et al. Citation1995). Hattori et al. (Citation2012) polymerized COs with DP from 6 to 15, thanks to lysozyme TG activity using CO3 as substrate. Similarly, Mallakuntla et al. (Citation2017) isolated a transglycosylating chitinase (EcChi1) from Enterobacter cloacae, able to synthesize short chain COs (with DP ranging from 4 to 9), starting from CO3 to CO6. In summary, enzymatic synthesis by transglycosylation appears as a very promising approach in terms of production costs, time and environmental impact, however, a great amount of purified GlcNAc monomers or oligomers is required as starting substrate.
As an alternative, chitinase hydrolytic activity can be exploited for chitin depolymerization. By expressing a recombinant chitinase from Chitinolyticabacter meiyuanensis in E. coli, Zhang et al. (Citation2018), were able to produce a considerable amount of CO2 and traces of CO3-4 from colloidal chitin. Similarly, a recombinant chitinase from Trichoderma harzianum expressed in Pichia pastoris led to the production of GlcNAc monomers and dimers from the digestion of colloidal chitin or chitosan (Kidibule et al. Citation2018). Lastly, Guan et al. (Citation2020) identified novel chitin deacetylase (CDA20) and chitosanase (CHIS5) enzymes from marine metagenome and successfully combined their use to obtain deacetylated Glc dimers and trimers.
Despite their major advantages, including mild reaction conditions, low pollution and time savings, enzymatic hydrolysis does not appear to be ideal for the large scale production of efficient Myc-factors, mainly because the reaction products are in most cases GlcNAc monomers, dimers or trimers, whereas the most active AM fungal signals are CO4 and CO5. Moreover, enzymatic techniques depend on the availability of high-quality enzymes, which have to be recovered after hydrolysis. This impacts on the production costs, limiting their application in industrial-scale production.
Biotechnological approaches
Genomic engineering of microbial cells (mainly bacteria) turned out to be a promising strategy to achieve higher CO yields for research and applicative uses. The design of optimized microbial strains by knockout, overexpression and transfer of target genes can be time and resource-consuming. Nevertheless it often grants high-yield and low-cost production of the desired molecules (Naqvi and Moerschbacher Citation2015). In Citation1995, Mergaert et al. expressed the nod gene cluster from Azorhizobium caulinodans in recombinant E. coli cells, obtaining different chitin oligomers and LCOs. Subsequently, Samain et al. (Citation1997) pushed this approach further, by expressing nodB and nodC in E. coli, and purified consistent amounts of mono-deacetylated CO5. More recently, Zhang et al. (Citation2007) cultured recombinant E. coli expressing nodC gene from Mesorhizobium loti. By using a two-step fermentation process and GlcNAc as a precursor, these authors obtained up to 930 mg/L of CO5 in a 10-L bioreactor. In Citation2019, Deng et al. blocked the expression of nagA (GlcNAc-6-phosphate deacetylase) and gamA (GlcN- 6-phosphate deaminase) in Corynebacterium glutamicum S9114, achieving a 54.8% increase in GlcNAc production and a final yield of 6.9 g/L GlcNAc in the culture broth.
In conclusion, such cell factory-based approaches have a number of advantages, compared to the previous production methods. This technique can provide pure COs and LCOs with the desired chemical features. Moreover, it does not require polluting solvents nor expensive purified enzymes, it is easily scalable and relatively affordable. Nevertheless, the biotechnological production of Myc-factors is still far from being industrially adopted. Additional research is needed to identify novel, more efficient chitin synthases besides rhizobial nod genes, possibly taking advantage of metagenomic approaches, before an environmentally-friendly industrial production of COs and LCOs can be able to satisfy the increasing demand for agricultural applications.
As a matter of fact, various formulations of chitin-derived products are being used in agriculture since decades, based on the empirical record of a general improvement in plant health, even if the precise mechanisms underpinning these positive effects had not been explained yet. As mentioned above, CO length has a major impact on the plant response they trigger. Mixed – often only partly characterized – formulations of COs have been shown to have antimicrobial and plant defense stimulation effects and are used for pest management. Xu et al. (Citation2007) demonstrated the inhibitory effect of short-chain deacetylated COs against in vitro cultured Phytophthora capsici. The same effect was confirmed by Nguyen et al. (Citation2018) using silver nanoparticles coated in short chitosan oligomers. COs antifungal activity was also investigated by Sun et al. (Citation2018), who used COs in combination with ϵ-poly-L-lysine on tomato plants, obtaining a significant protection against tomato gray mold infection. On the same line, foliar spray application of COs was reported to be effective in combating powdery mildew (Blumeria graminis) on barley (Faoro et al. Citation2008) and several additional examples can be found in literature, supporting the efficiency of CO treatment against a wide range of plant pathogens (Sarathchandra et al. Citation2004; Iriti et al. Citation2006; El Hadrami et al. Citation2010).
Most of these positive effects on plant health can be explained in the light of our current understanding of CO perception. The PAMP activity of longer COs is expected to trigger plant immunity, inducing a pre-alert status (or priming) prior to actual pathogen infection (Basa et al. Citation2020). In parallel, short-chain COs can reach the roots by rain elution and act as promoters of AM development. AM symbiosis is well known to contribute to raise plant defense in a priming-like condition; furthermore, a more extensive development of symbiosis improves plant nutrition and overall health and fitness, most likely allowing faster and more efficient defense responses in the case of a subsequent pathogen attack.
Besides their role in stimulating symbiosis and immunity, COs and LCOs have also been shown to promote plant growth, seed germination and photosynthesis in maize, rice, A. thaliana, cotton and wheat (Sun et al. Citation2015; Tanaka et al. Citation2015; Ané et al. Citation2016), while the induction of early flowering and an increased number of inflorescences was reported in orchids (Limpanavech et al. Citation2008). Such effects appear less directly related to either symbiosis – or defense-related signaling and stimulated further investigations. To this aim, Zhang et al. (Citation2016) used fully deacetylated chitooligomers of different length (DP2 to 12) on wheat seedlings, identifying CO7 as the most effective molecule in promoting biomass production and changing organic acid, sugar and amino acid composition within foliar tissues; by contrast CO2 and CO3 were completely ineffective. Comparable results were also obtained by Jia et al. (Citation2019) in soybean seedlings, where CO7 acted as a metabolic enhancer with the ability to modulate carbon and nitrogen metabolism.
Recently, COs were also found to improve plant resistance to abiotic stresses, such as drought, salinity and heavy metal toxicity (Zong et al. Citation2017; Zhou et al. Citation2018; Al-Ghamdi Citation2019; Muley et al. Citation2019; Turk et al. Citation2019). Such effects may be explained with the convergence of several plant responses to both biotic and abiotic stresses (Zhang et al. Citation2019), including the activation of ROS production and detoxification, cell wall modifications, proline and secondary metabolite accumulation. In other words, the priming effect caused by CO perception could also protect the plant in a context of abiotic stress (Zhang et al. Citation2019). Indeed, by treating salt-stressed wheat seedlings with COs of different length, Zhang et al. (Citation2017) showed that molecules with DP 6–8 (the best characterized chitin-derived PAMPs) were the most effective in stimulating antioxidant enzyme activity, photosynthesis and salt-related gene expression (SOS1 and NHX2).
Conclusions
One of the most striking aspects emerging from this rapid review of literature data is that the two major classes of AM signaling molecules – SLs and Myc-factors (mostly COs) – have been used as promoters of plant health in agriculture long before any of their signaling roles in symbiosis were elucidated. Given the evolutionary history of AM symbiosis, it is tempting to speculate that these molecules act in crucial processes that are deep-wired in the genomes of all extant descendants of those early land plants that first developed the ability to interact with beneficial soil fungi as they set to abandon the water environment and colonize land. As increasing evidence is accumulating in favor of the existence of parallel, largely unexplored, signaling processes between plants and AM fungi, research in the next few years is expected to bring to light more of such molecular switches and their multifaceted roles in plant development and interactions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors

Andrea Crosino
Andrea Crosino is a PhD student in Biology and Applied Biotechnologies at the University of Turin. Familiar with the concepts of circular economy, in particular in the field of biological waste/reuse, his research is focused on fungus-plant signaling in arbuscular mycorrhizal interactions and its manipulation for applicative outcomes.

Andrea Genre
Andrea Genre is Associate Professor in Plant Biology at the University of Turin. His research interests cover cellular and molecular aspects of arbuscular mycorrhizas, with a particular focus on fungal signals, plant signal transduction pathways, plant cell responses during fungal accommodation and agricultural applications of this symbiosis in a sustainable perspective.
References
- Abe S, et al. 2014. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci U S A 111:18084–18089. doi:10.1073/pnas.1410801111.
- Ajavakom A, et al. 2012. Products from microwave and ultrasonic wave assisted acid hydrolysis of chitin. Carbohydr Polym. 90:73–77. doi:10.1016/j.carbpol.2012.04.064.
- Akiyama K, et al. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 435:824–827.
- Akiyama K, et al. 2010. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51:1104–1117. doi:10.1093/pcp/pcq058.
- Al-Babili S, Bouwmeester H. 2015. Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186. doi:10.1146/annurev-arplant-043014-114759.
- Alder A, et al. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 335:1348–1351. doi:10.1126/science.1218094.
- Al-Ghamdi AA. 2019. Marjoram physiological and molecular performance under water stress and chitosan treatment. Acta Physiol Plant 41:44.
- Aliche EB, et al. 2020. Science and application of strigolactones. New Phytol 227:1001–1011. doi:10.1111/nph.16489.
- Alvira P, et al. 2016. Steam explosion for wheat straw pretreatment for sugars production. Bioethanol. 2:66–75. doi:10.1515/bioeth-2016-0003.
- Ané J-M, et al. 2002. Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol Plant-Microbe Interact 15(11):1108–1118.
- Ané J-M, et al. 2016. Chitin oligomers for use in promoting non-leguminous plant growth and development. US Patent 2016/0366883 A1.
- Arshad M, Frankenberger WT.. 2002. Ethylene in agriculture: synthetic and natural sources and applications. In: Arshad M, Frankenberger WT, editors. Ethylene. Boston: Springer; p. 290–294.
- Basa S, et al. 2020. The pattern of acetylation defines the priming activity of chitosan tetramers. J Am Chem Soc 142:1975–1986.
- Belmondo S, et al. 2017. Identification of genes involved in fungal responses to strigolactones using mutants from fungal pathogens. Curr Genet. 63:201–213. doi:10.1007/s00294-016-0626-y.
- Berlemont R. 2017. Distribution and diversity of enzymes for polysaccharide degradation in fungi. Sci Rep 7(1):222. doi:10.1038/s41598-017-00258-w.
- Besserer A, et al. 2006. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4:e226. doi:10.1371/journal.pbio.0040226.
- Besserer A, et al. 2008. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148:402–413. doi:10.1104/pp.108.121400.
- Bonfante & Requena. 2011. Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 14:451–457. doi:10.1016/j.pbi.2011.03.014.
- Bosso C, et al. 1986. The behavior of chitin towards anhydrous hydrogen fluoride. Preparation of b-(1-4)-linked 2- acetamido-2-deoxy-d-glucopyranosyl oligosaccharides. Carbohydr Res. 156:57–68.
- Boyer FD, et al. 2012. Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol 159:1524–1544. doi:10.1104/pp.112.195826.
- Boyer FD, et al. 2014. New strigolactone analogs as plant hormones with low activities in the rhizosphere. Mol. Plant. 7:675–690. doi:10.1093/mp/sst163.
- Boyno G, Demir S. 2022. Plant-mycorrhiza communication and mycorrhizae in inter-plant communication. Symbiosis. 86:155–168. doi:10.1007/s13199-022-00837-0.
- Brewer PB, et al. 2016. Lateral branching oxidoreductase acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc Natl Acad Sci U S A 113:6301–6306. doi:10.1073/pnas.1601729113.
- Chen ECH, et al. 2018. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 220:1161–1171. doi:10.1111/nph.14989.
- Chesterfield RJ, et al. 2020. Translation of strigolactones from plant hormone to agriculture: achievements, future perspectives, and challenges. Trends Plant Sci. 25(11):1087–1106. doi:10.1016/j.tplants.2020.06.005.
- Chevalier F, et al. 2014. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell. 26:1134–1150. doi:10.1105/tpc.114.122903.
- Chiu CH, Paszkowski U. 2021. How membrane receptors tread the fine balance between symbiosis and immunity signaling. PNAS. 118(24):e2106567118. doi:10.1073/pnas.2106567118.
- Choi J, et al. 2018. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annu Rev Phytopathol. 56:135–160. doi:10.1146/annurev-phyto-080516-035521.
- Choi J, et al. 2020. The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat Commun. 11:1114. doi:10.1038/s41467-020-16021-1.
- Conn CE, Nelson DC. 2016. Evidence that KARRIKIN-INSENSITIVE 2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6:1219. doi:10.3389/fpls.2015.01219.
- Cook CE, et al. 1966. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science. 154:1189–1190.
- Crosino A, et al. 2021. Extraction of short chain chitooligosaccharides from fungal biomass and their use as promoters of arbuscular mycorrhizal symbiosis. Sci Rep. 11:3798. doi:10.1038/s41598-021-83299-6.
- Defaye J, et al. 1994. A convenient access to beta-(1- 4)-linked 2-amino-2-deoxy-D-glucopyranosyl fluoride oligosaccharides and beta-(1-4)-linked 2-amino-2-deoxy-D-glucopyranosyl oligosaccharides by fluorolysis and fluorohydrolysis of chitosan. Carbohydr Res. 261:267–277.
- De Mesmaeker A, et al. 2019. Design, synthesis and biological evaluation of strigolactone and strigolactam derivatives for potential crop enhancement applications in modern agriculture. Chimia (Aarau). 73:549–560. doi:10.2533/chimia.2019.549.
- Denarie J, Cullimore J. 1993. Lipo-oligosaccharide nodulation factors: a new class of signaling molecules mediating recognition and morphogenesis. Cell. 74:951–954. doi:10.1016/0092-8674(93)90717-5.
- Dénarié J, Debellé F, Promé JC. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65:503–535.
- Deng C, et al. 2019. Metabolic engineering of Corynebacterium glutamicum S9114 based on whole-genome sequencing for efficient N-acetylglucosamine synthesis. Synthetic and Systems Biotechnology. 4:120–129.
- De Saint Germain A, et al. 2016. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12:787–794. doi:10.1038/nchembio.2147.
- Domard A, Carter C. 1989. Glucosamine oligomers: preparation and characterization. Int J Biol Macromol. 11:297–302.
- El Hadrami A, et al. 2010. Chitosan in plant protection. Mar Drugs. 8:968–987.
- Faoro F, et al. 2008. Chemical-induced resistance against powdery mildew in barley: the effects of chitosan and benzothiadiazole. BioControl. 53:387–401. doi:10.1007/s10526-007-9091-3.
- Feng F, et al. 2019. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat Commun. 10:5047. doi:10.1038/s41467-019-12999-5.
- Flematti GR, et al. 2015. What are karrikins and how were they ‘discovered’ by plants? BMC Biol. 13:108. doi:10.1186/s12915-015-0219-0.
- Foo E. 2013. Auxin influences strigolactones in pea mycorrhizal symbiosis. J Plant Physiol. 170:523–528. doi:10.1016/j.jplph.2012.11.002.
- Fridlender M, et al. 2015. Influx and efflux of strigolactones are actively regulated and involve the cell-trafficking system. Mol. Plant. 8:1809–1812. doi:10.1016/j.molp.2015.08.013.
- Fukui K, et al. 2013. Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol. Plant. 6:88–99. doi:10.1093/mp/sss138.
- Genre A, et al. 2013. Short chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198:190–202. doi:10.1111/nph.12146.
- Gobena D, et al. 2017. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes striga resistance. Proc Natl Acad Sci USA 114:4471–4476. doi:10.1073/pnas.1618965114.
- Gomez-Roldan V, et al. 2008. Strigolactone inhibition of shoot branching. Nature. 455:189–194.
- Grifoll-Romero L, et al. 2018. Chitin deacetylases: structures, specificities, and biotech applications. Polymers (Basel). 10(4):352. doi:10.3390/polym10040352.
- Groth M, et al. 2010. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell. 22:2509–2526. doi:10.1105/tpc.109.069807.
- Guan F, et al. 2020. Highly efficient production of chitooligosaccharides by enzymes mined directly from the marine metagenome. Carbohydr Polym. 234:115909. doi:10.1016/j.carbpol.2020.115909.
- Gutjahr C, et al. 2008. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 20(11):2989–3005. doi:10.1105/tpc.108.062414.
- Gutjahr C, et al. 2015. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science. 350:1521–1524. doi:10.1126/science.aac9715.
- Gutjahr C, Parniske M. 2013. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29:593–617. doi:10.1146/annurev-cellbio-101512-122413.
- Hamiaux C, et al. 2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22:2032–2036. doi:10.1016/j.cub.2012.08.007.
- Hao W. 2021. Review: advances in preparation of chitooligosaccharides with heterogeneous sequences and their bioactivity. Carb. Pol. 252:117206. doi:10.1016/j.carbpol.2020.117206.
- Hattori T, et al. 2012. Enzymatic synthesis of an a-chitin-like substance via lysozyme-mediated transglycosylation. Carbohydr Res 347:16–22. doi:10.1016/j.carres.2011.09.025.
- Hauck C, et al. 1992. A germination stimulant for parasitic flowering plants from Sorghum bicolor, a genuine host plant. J Plant Physiol 139:474–478.
- He J, et al. 2019. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol Plant. 12:1561–1576. doi:10.1016/j.molp.2019.10.015.
- Imaizumi-Anraku H, et al. 2005. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature. 433(7025):527–531. doi:10.1038/nature03237.
- Iriti M, et al. 2006. Cell death-mediated antiviral effect of chitosan in tobacco. Plant Physiol Biochem. 44:893–900.
- Jia Y, et al. 2019. Effects of different oligochitosans on isoflavone metabolites, antioxidant activity, and isoflavone biosynthetic genes in soybean (Glycine max) seeds during germination. J Agric Food Chem 67:4652–4661.
- Jin Y, et al. 2016. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat Commun. 7:12433. doi:10.1038/ncomms12433.
- Kanamori N, et al. 2006. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proceedings of the National Academy of Sciences, USA. 103:359–364. doi:10.1073/pnas.0508883103.
- Kang B, et al. 2007. Synergetic degradation of chitosan with gamma radiation and hydrogen peroxide. Polym Degrad Stab. 92:359–362.
- Kapulnik, Y. et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216 (2011). doi:10.1007/s00425-010-1310-y
- Kasprzewska A. 2003. Plant chitinases—regulation and function. Cell Mol Biol Lett 8(3):809–824.
- Kazami N, et al. 2015. A simple procedure for preparing chitin oligomers through acetone precipitation after hydrolysis in concentrated hydrochloric acid. Carbohyd. Polym. 132:304–310. doi:10.1016/j.carbpol.2015.05.082.
- Kevei Z, et al. 2007. 3-Hydroxy-3- methylglutaryl coenzyme A reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell. 19:3974–3989. doi:10.1105/tpc.107.053975.
- Khokhani D, et al. 2021. Deciphering the chitin code in plant symbiosis, defense, and microbial networks. Annu Rev Microbiol. 75:583–607. doi:10.1146/annurev-micro-051921-114809.
- Kidibule PE, et al. 2018. Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity. Microb. Cell. Fact. 17:47. doi:10.1186/s12934-018-0895-x.
- Kobae Y. 2018. Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 59:544–553. doi:10.1093/pcp/pcy001.
- Kohlen W, et al. 2011. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155:974–987. doi:10.1104/pp.110.164640.
- Kountche BA, et al. 2019. Suicidal germination as a control strategy for Striga hermonthica (Benth.) in smallholder farms of sub-saharan Africa. Plants People Planet. 1:107–118. doi:10.1002/ppp3.32.
- Lanfranco L, et al. 2018. Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J Exp Bot. 69:2175–2188. doi:10.1093/jxb/erx432.
- Lanfranco L, Bonfante P, Genre A. 2016. The mutualistic interaction between plants and arbuscular mycorrhizal fungi. Microbiol. Spect. 4:727–747. doi:10.1128/microbiolspec.funk-0012-2016.
- Langner T, Göhre V. 2016. Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Curr Genet 62(2):243–254. doi:10.1007/s00294-015-0530-x.
- Lee CG, et al. 2008. Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol 20(6):684–689. doi:10.1016/j.coi.2008.10.002.
- Lee HW, et al. 2020. Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. J Biol Chem 295:4181–4193. doi:10.1074/jbc.RA119.011509.
- Liao D, et al. 2018. Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front Plant Sci. 9:1004. doi:10.3389/fpls.2018.01004.
- Limpanavech P, et al. 2008. Chitosan effects on floral production, gene expression, and anatomical changes in the Dendrobium orchid. Sci Hortic 116:65–72.
- Ling M, et al. 2018. Metabolic engineering for the production of chitooligosaccharides: advances and perspectives. Emerg. Top. Life Sci. 2:377–388. doi:10.1042/ETLS20180009.
- Lodhi, G., et al. 2014. Chitooligosaccharide and Its derivatives: preparation and biological applications. BioMed Res Int, ID 654913. doi:10.1155/2014/654913
- Machová E, et al. 1999. Effect of ultrasonic treatment on the molecular weight of carboxymethylated chitin–glucan complex from Aspergillus niger. Ultrason Sonochem. 5:169–172.
- Maillet F, et al. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 469:58–63.
- Mallakuntla MK, et al. 2017. Transglycosylation by a chitinase from Enterobacter cloacae subsp. cloacae generates longer chitin oligosaccharides. Sci Rep 7:1–12. doi:10.1038/s41598-017-05140-3.
- Mergaert P, et al. 1995. Biosynthesis of Azorhizobium caulinodans nod factors. J Biol Chem. 270:29217–29223.
- Merzendorfer H. 2011. The cellular basis of chitin synthesis in fungi and insects: common principles and differences. Eur J Cell Biol 90(9):759–769. doi:10.1016/j.ejcb.2011.04.014.
- Miyakawa T, et al. 2019. Molecular basis of strigolactone perception in root-parasitic plants: aiming to control its germination with strigolactone agonists/antagonists. Cell Mol Life Sci. 77:1103–1113. doi:10.1007/s00018-019-03318-8.
- Mori N, et al. 2020. Identification of two oxygenase genes involved in the respective biosynthetic pathways of canonical and non-canonical strigolactones in Lotus japonicus. Planta. 251:40. doi:10.1007/s00425-019-03332-x.
- Moscatiello R, et al. 2014. The intracellular delivery of TAT-aequorin reveals calcium mediated sensing of environmental and symbiotic signals by the arbuscular mycorrhizal fungus Gigaspora margarita. New Phytol. 203:1012–1020. doi:10.1111/nph.12849.
- Mourya VK, et al. 2011. Chitooligosaccharides: synthesis, characterization and applications. Polym. Sci. Ser. A. 53:583–612. doi:10.1134/S0965545X11070066.
- Mukherjee A, Ané J-M. 2011. Germinating spore exudates from arbuscular mycorrhizal fungi: molecular and developmental responses in plants and their regulation by ethylene. Mol Plant-Microbe Interact 24(2):260–270. doi:10.1094/MPMI-06-10-0146.
- Muley AB, et al. 2019. Gamma radiation degradation of chitosan for application in growth promotion and induction of stress tolerance in potato (Solanum tuberosum L. Carbohydr Polym 210:289–301.
- Müller S, et al. 1992. Germination stimulants produced by Vigna unguiculata Walp cv Saunders upright. J Plant Growth Regul 11:77–84.
- Nakamura H, et al. 2013. Molecular mechanismof strigolactone perception by DWARF14. Nat Commun 4:2613. doi:10.1038/ncomms3613.
- Naqvi S, Moerschbacher BM. 2015. The cell factory approach toward biotechnological production of high-value chitosan oligomers and their derivatives: an update. Crit Rev Biotechnol. doi:10.3109/07388551.2015.1104289.
- Nasir F, et al. 2018. Current understanding of pattern-triggered immunity and hormone-mediated defense in rice (Oryza sativa) in response to Magnaporthe oryzae infection. Semin Cell Dev Biol. 83:95–105. doi:10.1016/j.semcdb2017.10.020.
- Nefkens GHL, et al. 1997. Synthesis of a phthaloylglycine-derived strigol analogue and its germination stimulatory activity toward seeds of the parasitic weeds Striga hermonthica and Orobanche crenata. Journal of Agriculture and Food Chemistry. 45:2273–2277.
- Nguyen VT, et al. 2018. Effect of oligochitosan-coated silver nanoparticles (OCAgNPs) on the growth and reproduction of three species Phytophthora in vitro. Arch. Phytopathol. Plant Prot. 51:227–240.
- Niu YF, et al. 2013. Responses of root architecture development to low phosphorus availability: a review. Ann Bot 112:391–408. doi:10.1093/aob/mcs285.
- Oldroyd GED. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11:252–263. doi:10.1038/nrmicro2990.
- Orlean P, Funai D. 2019. Priming and elongation of chitin chains: implications for chitin synthase mechanism. The Cell Surface. 5:100017. doi:10.1016/j.tcsw.2018.100017.
- Osada M, et al. 2013. Effects of supercritical water and mechanochemical grinding treatments on physicochemical properties of chitin. Carbohydr Polym. 92:1573–1578. doi:10.1016/j.carbpol.2012.10.068.
- Parker C. 2009. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci 65:453–459. doi:10.1002/ps.1713.
- Passos H, et al. 2014. Ionic liquid solutions as extractive solvents for value-added compounds from biomass. Green Chem 16:4786–4815. doi:10.1039/C4GC00236A.
- Peiter E, et al. 2007. The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol 145(1):192–203. doi:10.1104/pp.107.097261.
- Pimprikar P, et al. 2016. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol. 26:987–998. doi:10.1016/j.cub.2016.01.069.
- Poinsot V, et al. 2016. New insights into Nod factor biosynthesis: analyses of chitooligomers and lipo-chitooligomers of Rhizobium sp. IRBG74 mutants. Carbohydr Res 434:83–93. doi:10.1016/j.carres.2016.08.001.
- Prandi C, et al. 2014. Tailoring fluorescent strigolactones for in vivo investigations: a computational and experimental study. Org Biomol Chem 12:2960–2968. doi:10.1039/C3OB42592D.
- Rasmussen A, et al. 2013. A fluorescent alternative to the synthetic strigolactone GR24. Mol. Plant. 6:100–112. doi:10.1093/mp/sss110.
- Rich MK, Nouri E, Courty PE, Reinhardt D. 2017. Diet of arbuscular mycorrhizal fungi: bread and butter? Trends Plant Sc. 22:652–660.
- Rodenburg J. 2016. Parasitic weed incidence and related economic losses in rice in Africa. Agric Ecosyst Environ. 235:306–317. doi:10.1016/j.agee.2016.10.020.
- Roy I, et al. 2003. Accelerating enzymatic hydrolysis of chitin by microwave pretreatment. Biotechnol Prog. 19:1648–1653.
- Rush TA, et al. 2020. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun 11(1):3897. doi:10.1038/s41467-020-17615-5.
- Saito K, et al. 2007. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell. 19:610–624. doi:10.1105/tpc.106.046938.
- Samain E, et al. 1997. Gram-scale synthesis of recombinant chitooligosaccharides in Escherichia coli. Carbohydr Res. 302:35–42.
- Samejima H, et al. 2016. Practicality of the suicidal germination approach for controlling Striga hermonthica. Pest Manag Sci 72:2035–2042. doi:10.1002/ps.4215.
- Sarathchandra RG, et al. 2004. A chitosan formulation ElexaTM induces downy mildew disease resistance and growth promotion in pearl millet. Crop Prot. 23:881–888.
- Savage PE. 1999. Organic chemical reactions in supercritical water. Chem Rev. 99:603–622.
- Scaffidi A, et al. 2014. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165:1221–1232. doi:10.1104/pp.114.240036.
- Seto Y, et al. 2019. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10:191. doi:10.1038/s41467-018-08124-7.
- Shabek N, et al. 2018. Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature. 563:652–656. doi:10.1038/s41586-018-0743-5.
- Shahul Hameed U, et al. 2018. Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor. EMBO Rep 19:e45619. doi:10.15252/embr.201745619.
- Shimizu T, et al. 2010. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64:204–214. doi:10.1111/j.1365-313X.2010.04324.x.
- Siame BA, et al. 1993. Isolation of strigol, a germination stimulant for Striga asiatica, from host plants. J Agric Food Chem 41:1486–1491.
- Singh S, et al. 1995. Glycosidase-catalysed oligosaccharide synthesis: preparation of N-acetylchitooligosaccharides using the b-N-acetylhexosaminidase of Aspergillus oryzae. Carbohydr Res Res. 279:293–305.
- Stanga JP, et al. 2016. Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta. 243:1397–1406. doi:10.1007/s00425-015-2458-2.
- Sun G, et al. 2018. Synergistic effect of the combined bio-fungicides “-poly-L-lysine and chitooligosaccharide in controlling grey mould (Botrytis cinerea) in tomatoes. Int. J. Food Microbiol. 276:46–53.
- Sun J, et al. 2015. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 27:823–838. doi:10.1105/tpc.114.131326.
- Takahashi Y, et al. 1995. Effect of sonolysis on acid degradation of chitin to form oligosaccharides. Bull Chem Soc Jpn. 68:851–857.
- Tanaka K, et al. 2015. Effect of lipo-chitooligosaccharide on early growth of C4 grass seedlings. J Exp Bot 66(19):5727–5738.
- Tian F, et al. 2003. The depolymerization mechanism of chitosan by hydrogen peroxide. J Mater Sci. 38:4709–4712.
- Tømmeraas K, et al. 2001. Preparation and characterisation of oligosaccharides produced by nitrous acid depolymerisation of chitosans. Carbohydr Res. 333:137–144.
- Tsuchiya Y, et al. 2015. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science. 349:864–868. doi:10.1126/science.aab3831.
- Tsuzuki S, et al. 2016. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol Plant-Microbe Interact 29:277–286. doi:10.1094/MPMI-10-15-0234-R.
- Turk H, et al. 2019. Chitosan-induced enhanced expression and activation of alternative oxidase confer tolerance to salt stress in maize seedlings. Plant Physiol Biochem 141:415–422.
- Ueno K, et al. 2014. Heliolactone, a nonsesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry. 108:122–128. doi:10.1016/j.phytochem.2014.09.018.
- Umehara M, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 455:195–200.
- Umehara M, et al. 2010. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51:1118–1126. doi:10.1093/pcp/pcq084.
- Uraguchi D, et al. 2018. A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science. 362:1301–1305. doi:10.1126/science.aau5445.
- Vassilev N, et al. 2015. Unexploited potential of some biotechnological techniques for biofertilizer production and formulation. Appl Microbiol Biotechnol. 99(12):4983–4996. doi:10.1007/s00253-015-6656-4.
- Vekariya RL. 2017. A review of ionic liquids: applications towards catalytic organic transformations. J Mol Liq 227:44–60. doi:10.1016/j.molliq.2016.11.123.
- Venkateshwaran M, et al. 2012. The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell. 24(6):2528–2545. doi:10.1105/tpc.112.098475.
- Venkateshwaran M, et al. 2015. A role for the mevalonate pathway in early plant symbiotic signaling. Proceedings of the National Academy of Sciences, USA. 112:9781–9786. doi:10.1073/pnas.1413762112.
- Vickers CE, et al. 2015. Production of industrially relevant isoprenoid compounds. In: Kamm B, editor. Engineered microbes. Springer; p. 303–334.
- Volk H, et al. 2019. Chitin-binding protein of Verticillium nonalfalfae disguises fungus from plant chitinases and suppresses chitin-triggered host immunity. Mol Plant-Microbe Interact 32(10):1378–1390. doi:10.1094/MPMI-03-19-0079-R.
- Volpe V, et al. 2020. Short chain chito-oligosaccharides promote arbuscular mycorrhizal colonization in Medicago truncatula. Carbohydr Polym 229:115505. doi:10.1016/j.carbpol.2019.115505.
- Wakabayashi T, et al. 2019. Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Sci Adv 5:eaax9067. doi:10.1126/sciadv.aax9067.
- Wakabayashi T, et al. 2020. CYP722C from Gossypium arboreum catalyzes the conversion of carlactonoic acid to 5-deoxystrigol. Planta. 251(5):97. doi:10.1007/s00425-020-03390-6.
- Wakabayashi T, et al. 2021. Identification and characterization of sorgomol synthase in sorghum strigolactone biosynthesis. Plant. Phys. 185(3):902–913. doi:10.1093/plphys/kiaa113.
- Walker CH, et al. 2019. Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol 17:70. doi:10.1186/s12915-019-0689-6.
- Wan J, et al. 2008. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 20(2):471–481. doi:10.1105/tpc.107.056754.
- Wilkes JS. 2004. Properties of ionic liquid solvents for catalysis. J.Mol. Catal. A. 214:11–17. doi:10.1016/j.molcata.2003.11.029.
- Wu HS, et al. 2011. Process for producing glucosamine and acetyl glucosamine by microwave technique. US Patent 20110114472 A1.
- Wu S. 2021. Establishment of strigolactone-producing bacterium-yeast consortium. Sci. Adv. 7:eabh4048. doi:10.1126/sciadv.abh4048.
- Xie X, et al. 2013. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant. 6:153–163.
- Xu J, et al. 2007. Antifungal activity of oligochitosan against Phytophthora capsici and other plant pathogenic fungi in vitro. Pestic Biochem Physiol 87:220–228.
- Yamago S, et al. 2004. Iterative glycosylation of 2-deoxy-2-aminothioglycosides and its application to the combinatorial synthesis of linear oligoglucosamines. Angew Chem. 116(16):2197–2200.
- Yang C, Wang E, Liu J. 2022. CERK1, more than a co-receptor in plant–microbe interactions. New Phytol 234:1606–1613. doi:10.1111/nph.18074.
- Yao R, et al. 2016. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature. 536:469–473. doi:10.1038/nature19073.
- Yokota T, et al. 1998. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry. 49:1967–1973.
- Yoneyama K, et al. 2007. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta. 227:125–132.
- Yoneyama K, et al. 2010. Strigolactones as germination stimulants for root parasitic plants. Plant Cell Physiol 51:1095–1103. doi:10.1093/pcp/pcq055.
- Yoneyama K, et al. 2013. Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta. 238:885–894. doi:10.1007/s00425-013-1943-8.
- Yoneyama K, et al. 2015. Difference in striga-susceptibility is reflected in strigolactone secretion profile, but not in compatibility and host preference in arbuscular mycorrhizal symbiosis in two maize cultivars. New Phytol 206:983–989. doi:10.1111/nph.13375.
- Yoneyama K, et al. 2018. Which are the major players, canonical or non-canonical strigolactones? J Exp Bot 69:2231–2239. doi:10.1093/jxb/ery090.
- Yoneyama K, et al. 2020. Hydroxyl carlactone derivatives are predominant strigolactones in Arabidopsis. Plant Direct. 4:5. doi:10.1002/pld3.219.
- Yoneyama K, Brewer PB. 2021. Strigolactones, how are they synthesized to regulate plant growth and development? Curr Opin Plant Biol. 63:102072. doi:10.1016/j.pbi.2021.102072.
- Yoshida S, et al. 2012. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol 196:1208–1216. doi:10.1111/j.1469-8137.2012.04339.x.
- Yu N, et al. 2014. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 24:130–133. doi:10.1038/cr.2013.167.
- Zhang A, et al. 2018. Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-d-glucosamine production. Biotechnol Biofuels. 11:179. doi:10.1186/s13068-018-1169-x.
- Zhang D, et al. 2007. A two-step fermentation process for efficient production of penta-N-acetyl-chitopentaose in recombinant Escherichia coli. Biotechnol Lett. 29:1729–1733. doi:10.1007/s10529-007-9462-y.
- Zhang X, et al. 2016. Size effects of chitooligomers on the growth and photosynthetic characteristics of wheat seedlings. Carbohydr Polym 138:27–33.
- Zhang X, et al. 2017. Relationship between the degree of polymerization of chitooligomers and their activity affecting the growth of wheat seedlings under salt stress. J. Agric. Food Chem. 65:501–509.
- Zhang X, et al. 2021. Efficient production of oligomeric chitin with narrow distributions of degree of polymerization using sonication-assisted phosphoric acid hydrolysis. Carbohydr Polym. 276:118736. doi:10.1016/j.carbpol.2021.118736.
- Zhang Y, et al. 2014. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol 10:1028–1033. doi:10.1016/j.jpowsour.2013.09.135.
- Zhang Y, et al. 2019. Chitooligosaccharide plays essential roles in regulating proline metabolism and cold stress tolerance in rice seedlings. Acta Physiol Plant 41:77.
- Zhao LH, et al. 2013. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23:436–439. doi:10.1038/cr.2013.19.
- Zhou F, et al. 2013. D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature. 504:406–410. doi:10.1038/nature12878.
- Zhou J, et al. 2018. Chitooligosaccharides enhance cold tolerance by repairing photodamaged PS II in rice. J Agric Sci 156:888–899.
- Zipfel C, Oldroyd GE. 2017. Plant signalling in symbiosis and immunity. Nature. 543:328–336. doi:10.1038/nature22009.
- Zong H, et al. 2017. Improvement in cadmium tolerance of edible rape (Brassica rapa L.) with exogenous application of chitooligosaccharide. Chemosphere. 181:92–100.
- Zwanenburg B, et al. 2016. Suicidal germination for parasitic weed control. Pest Manag Sci 72:2016–2025. doi:10.1002/ps.4222.
- Zwanenburg B, Pospíšil T. 2013. Structure and activity of strigolactones: new plant hormones with a rich future. Mol Plant. 6:38–62. doi:10.1093/mp/sss141.
