ABSTRACT
The use of biological control agents (BCAs) to cope with diseases has received considerable attention owing to its high efficiency and environmental safety. The aim of our study was to investigate the potential role of Pseudomonas mediterranea pretreatment in the response of lemon [Citrus limon (L.) Burm. f.] against mal secco, which is a devastating citrus disease caused by the fungus Plenodomus tracheiphilus. RNAseq analysis revealed that the fungus induced marked reprogramming of the transcriptome, but P. mediterranea pretreatment strongly reduced lemon leaf transcriptome modifications and limited the amount of fungal DNA inside the plant tissue. Furthermore, P. mediterranea prevented the downregulation of the genes involved in effector-triggered immunity (ETI) and the deregulation of genes involved in the biosynthesis and perception of the main phytohormones. To the best of our knowledge, this work represents the first report on the analysis of the P. tracheiphilus-lemon plant-BCA interaction at the molecular level.
1. Introduction
Citrus is an economically important fruit crop that is seriously afflicted by several biotic stresses caused by viruses, bacteria and fungi that limit the quantity and quality of production (Talon and Gmitter Citation2008). The lemon industry in the Mediterranean basin is strongly threatened by ‘mal secco' disease (MSD), a serious vascular disease caused by the mitosporic fungus Plenodomus tracheiphilus. The pathogen mainly infects lemon [C. limon (L.) Burm. F. ], citron (C. medica L.) and other citrus species and hybrids that have citron or lemon as a parent (Nigro et al. Citation2011; Catara and Catara Citation2019; Russo et al. Citation2020, Citation2021). The disease’s symptoms include the desiccation of twigs, branches and the entire plant; hence, the term ‘mal secco' is used to specify the disease worldwide. The disease typically begins to manifest in the spring on the leaves of the upper shoots, which initially exhibit a mild discolouration of the main and secondary veins before turning yellow or occasionally brown and dropping to the ground (Migheli et al. Citation2009; Batuman et al. Citation2020). Afterwards, progressive basipetal desiccation occurs, and finally, the entire plant dies. Currently, it can be estimated that the pressure of disease has led to a reduction of more than 80 percent of lemon crop acreage in the last 30 years with economic losses in terms of jobs and industry output (Pérez-Tornero et al. Citation2012). The exploitation of genetic tolerance with the use of less susceptible varieties and rootstocks for new plantings is certainly a viable strategy for containing the disease. However, to date, traditional breeding programs and biotechnological techniques have not yet led to the development of new varieties that combine mal secco tolerance with excellent fruit quality (Gentile and La Malfa Citation2019; Poles et al. Citation2020; Catalano et al. Citation2021). The management of MDS is therefore based on the adoption of preventive measures such as the use of healthy plants from certified nurseries or spray applications of copper-based compounds either after pruning or other natural events that cause wounds (hailstorm, frost) from which the fungus can penetrate (Deng et al. Citation2009). However, the use of antimicrobial pesticides is mainly discouraged by international regulations on permitted levels of chemical residues. In particular, the amount of copper allowed in plant protection has been restricted over time by EU legislation, and the final objective would be to phase copper out (Tamm et al. Citation2022). Thus, there is a critical need for novel, efficient control tools that can supplement or even replace the use of chemical antimicrobials in agricultural production to guarantee high production standards and greater sustainability. Among natural products, beneficial microorganisms (biocontrol and/or plant biostimulant microorganisms) seem to be the most promising tools for guaranteeing plant health, as well as the quality and safety of plant products (Palmieri et al. Citation2022; Kelbessa et al. Citation2023). A biological control agent (BCA) is a microorganism capable of counteracting one or more target plant pathogens, interfering with their life cycles (Corke Citation1974). Different modes of action through which biocontrol agents hinder the spread of plant pathogens have been identified, such as the occurrence either of direct antagonism or by inducing resistance in plants, exercising indirect biocontrol activity (Raymaekers et al. Citation2020). In recent decades, research activity has focused on evaluating the efficacy of many selected BCAs against various harmful fungal pathogens, BCA-based bioformulations are now available on the market, and others are waiting for final approval by the authorities (Ghadamgahi et al. Citation2022; Lahlali et al. Citation2022). Recently, the effectiveness of biological commercial products based on Bacillus amyloliquefaciens against Plenodomus tracheiphilus was evaluated through in vitro and in vivo experiments (Aiello et al. Citation2022). Regarding the in planta experiment, it was observed that biological products can significantly reduce disease incidence and symptom severity in pretreated C. volkameriana seedlings (Aiello et al. Citation2022). However, the detailed citrus plant response to beneficial agents and the possible protective effects against P. tracheiphilus have not yet been investigated at the molecular level.
The now consolidated exploitation of high-performance sequencing platforms to analyse the regulatory mechanisms of plants under specific conditions represents the basis for the development of innovative tools to minimize or replace the massive use of chemical antimicrobials. Hence, RNA sequencing (RNA-seq), with the use of next-generation sequencing techniques (NGS), offers the opportunity to isolate genes of interest, develop functional markers, and quantify gene expression (Sicilia et al. Citation2019; Sicilia et al. Citation2020). In addition, it provides an unprecedented high-resolution view of the transcriptional profile of the host when infected with a pathogen (Russo et al. Citation2021) and of the same pathogen during in planta infection (Sicilia et al. Citation2022). Recent studies concerning the transcriptomic response of rough lemon (Citrus jambhiri Lush), which is particularly susceptible to the disease, inoculated with P. tracheiphilus highlighted the triggering of an acute response, indicated by a total of 4986 differentially expressed genes (DEGs), attributable to the transduction pathway induced by the synthesis of the phytohormone salicylic acid (Russo et al. Citation2021). The results of this work indicate that fungal infection represses the reactions of chlorophyll photosynthesis and iron absorption processes and is responsible both for a lack of energy needed for plants to carry out vital functions and for the chlorosis found on the adaxial face of leaves (Russo et al. Citation2021). Furthermore, the genes involved in the hypersensitive response (HR) (Jones and Dangl Citation2006; Chang et al. Citation2019), which would lead to the confinement of the pathogen, are repressed in plants inoculated with the fungus, which can expand inside the plant (Russo et al. Citation2021). Pseudomonas spp. produce antifungal metabolites representing a real alternative to the application of chemical fungicides (Walsh et al. Citation2001). Bacterial strains belonging to the Pseudomonas spp. have been isolated mainly from the root environment and from various plant matrices (Catara et al. Citation2002; Catara Citation2007; Strano et al. Citation2017). These strains have shown high antagonistic activity against phytopathogenic bacteria, oomycetes and fungi due to the production of a plethora of secondary metabolites and have been proposed as biological control agents (Catara Citation2007; Strano et al. Citation2017). In particular, promising results have been preliminary obtained using P. mediterranea 3C that significantly reduced fungal infection when injected in the leaf midvein of sour orange 48 h before the challenge inoculation (Coco et al. Citation2004).
Considering the importance of gaining new insights into plant defence primed by BCAs at molecular level and in view of an eco-sustainable strategy against mal secco disease, the aim of this work was to investigate the effect of the application of cell suspensions of P. mediterranea on the transcriptome of ‘Femminello Siracusano 2Kr’ lemon leaves [C. limon (L.) Burm. F.] artificially inoculated with P. tracheiphilus during the initial, asymptomatic phase of the disease, using NGS (RNA-seq) and de novo RNA assembly approaches.
2. Materials and methods
2.1. Preparation of Pseudomonas mediterranea cell-based bioformulation
The bacterial inoculum was obtained from cultures of P. mediterranea strain PVCT 3C grown overnight at 28°C in the liquid substrate Luria Broth (LB) under stirring conditions (180 rpm). Bacterial cells were harvested by centrifugation at 7500 rpm for 15 min. The pellet was suspended in sterile water up to a concentration of 108 cfu ml−1 equal to an optical density (OD) of OD600 = 1.0 measured by a spectrophotometer at a wavelength of 600 nm. Plants were treated by aerial spraying of the foliage with the bacterium suspension (see paragraph 2.3), both at three days and one day before fungal pathogen inoculation.
2.2. Preparation of inoculum of the fungal pathogen
The inoculum of P. tracheiphilus consisted of vial phialoconidia of the PVCT Pt57 strain obtained in carrot broth (Dimaria et al. Citation2023). Briefly, two PDA plugs taken along the growing margins of 7-day-old fungal colonies were transferred into 250 mL breakwater indented flasks containing 150 mL of carrot broth. The flasks were incubated under continuous stirring (220 rpm) for four days at 22°C under diffuse light conditions. Conidia were obtained by centrifugation (6000 rpm for 15 min) of the culture broth previously filtered to remove the mycelial component. Phialoconidia were suspended in dH2O at a concentration of 106 ml−1.
2.3. Experimental scheme
Potted lemon ‘Femminello Siracusano 2Kr’ plants grafted onto Citrus aurantium (sour orange) kept in greenhouses at Agrobiotech srl (Catania) were used for the inoculation tests. The inoculations with the fungal pathogen were carried out by placing 10 µl of the vial suspension of P. tracheiphilus isolated PVCT Pt57 (see paragraph 2.2) on the secondary veins of young but fully expanded leaves; the droplets were then pierced until the leaf was incised with an awl with three entomological needles mounted at the end (Oliveri et al. Citation2022). Each leaf was inoculated in six spots. To evaluate the effect of the treatment with the biocontrol agent, the following theses were defined as follows: 3C thesis, plants treated with the biocontrol agent three days and one day before H2O inoculation; 3CPt thesis, plants treated with the biocontrol agent three days and one day before P. tracheiphilus inoculation; Pt thesis: plants treated with water three days and one day before P. tracheiphilus inoculation; and CK thesis, plants treated and inoculated with dH2O. Six plants were inoculated for each thesis and, in each of these, three branches and at least six leaves per branch, for a total of approximately 18 leaves per plant. The leaves intended for transcriptomic analysis were collected 7 days after inoculation before symptom appearance (Figure S1).
2.4. DNA extraction and confirmation of infection by real-time PCR
On the seventh day after inoculation, three leaves were taken for each thesis (washed with RNase-free H2O to eliminate the excess bacteria on the surface). The collected leaves were immediately frozen in situ with liquid nitrogen and stored at −80°C until DNA extraction. DNA extraction was performed with the CTAB protocol (Springer Citation2010) after powdering the frozen samples with a mortar and pestle using liquid nitrogen. DNA concentration and purity were checked using a Nanodrop 2000 spectrophotometer (Thermo Scientific™, Waltham, MA, USA). The real-time PCR assay was performed with the method described by (Licciardello et al. Citation2006) modified by Russo et al. (Citation2011). Each reaction was performed in a final volume of 15 µl with Taq Quanti Nova mix (Probe PCR kit- 500, Qiagen). In addition to the master mix at the concentration indicated by the company, the reaction included 400 nM of each of the two primers, GR70 (5ʹ-GTACCGTACGCCTTGGGGAC-3ʹ) and GL1 (5ʹ- GAAGCGTTTGGAGGAGAGAATG-3ʹ), 200 nM of the PP1-labelled probe (5ʹ-FAM-CAACGCAATCTTGGCGACTGTCGTT- BHQ1-3ʹ), and 2 µl of fungal genomic DNA corresponding to the positive control or, in the case of the negative control, 2 µl of sterile water (Licciardello et al. Citation2006). Real-time PCRs were performed with the BioRAD iCyclerIQ instrument. The reaction conditions were as follows: an initial denaturation at 95°C for 2 min (1 cycle) followed by 40 cycles at 95°C for 10 s, 62°C for 15 s and 72°C for 15 s. For the absolute quantification of fungal DNA, a real-time PCR standard curve was constructed using serial dilutions in sterile distilled water of the isolated PVCT DNA Pt57 (100 µg ml−1). The standard curve was obtained by graphing the Ct values corresponding to the logarithmic concentration of each serial dilution. Ct values were calculated for each standard from three PCR replicates performed in three separate runs.
2.5. Sample collection and RNA extraction
On the seventh day after inoculation, three leaves were taken for each thesis (washed with RNase-free H2O to eliminate the excess bacteria on the surface). The collected leaves were immediately frozen in liquid nitrogen in situ and stored at −80°C. RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions as detailed in (Russo et al. Citation2021).
2.6. Preparation of libraries and RNA sequencing
After RNA quantification and quality control procedures (Russo et al. Citation2021), sequencing libraries were generated as reported in (Sicilia et al. Citation2019; Russo et al. Citation2021).
2.7. De novo assembly of the transcriptome and functional annotation
De novo assembly of the transcriptome was performed using Trinity (version r20140413p1) with parameters min_kmer_cov:2 (k = 25). Next, hierarchical clustering was performed with Corset (version v1.05, https://github.com/Oshlack/Corset/wiki) to remove redundancies (parameter -m 10), and the longest transcripts of each cluster were selected as unigenes. The Citrus limon transcriptome was submitted to NCBI (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE227934. Gene functions were annotated using the following databases: National Center for Biotechnology Information (NCBI), nonredundant protein sequences (Nr), NCBI nonredundant nucleotide sequences (Nt), family of proteins (Pfam), Clusters of Orthologous Groups of proteins (KOG/COG), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), Ortholog database (KO) and Gene Ontology (GO). Pathway analysis was conducted using MapMan3.6.0RC1 (https://mapman.gabipd.org/, accessed 19 September 2022). All unigenes were annotated and mapped using Mercator4 V2.0, an online tool by MapMan (https://www.plabipd.de/portal/mercator4, visited 20 September 2022), which accurately assigns hierarchical ontology. The significant DEGs (padj ≤ 0.05), with the corresponding log2FoldChange values, were used as the dataset to be aligned to the Mercator map.
2.8. Quantification of gene expression and analysis of differential expression
Gene expression levels were estimated via RSEM (version v1.2.26, http://deweylab.github.io/RSEM/) with bowtie2 mismatch 0 parameters to map the Corset-filtered transcriptome. For each sample, the clean reads were mapped back onto the assembled transcriptome, and the number of reads for each gene was obtained. Differential expression analysis was performed using the R DESeq package (version 1.12.0, padj ≤ 0.05, https://bioconductor.org/packages/release/bioc/html/DESeq.html) considering the following comparisons: Pt vs. CK (fungus inoculated vs. control), 3C vs. CK (bacterial sprayed vs. control) and 3CPt vs. CK (bacterial sprayed and fungus inoculated vs. control). The resulting padj values were adjusted using the Benjamini and Hochberg approach to control for the false discovery rate (FDR) (Benjamini and Hochberg Citation1995). Genes with a padj value ≤ 0.05 were assigned as differentially expressed. A log2FoldChange threshold of 0.58 (1.5-fold change) was adopted. GO enrichment analysis of differentially expressed genes (DEGs) was implemented by GOseq R packages (version 1.10.0, 2.10.0, corrected p value ≤ 0.05 based on Wallenius noncentral hypergeometric distribution). In addition, all unigenes underwent systematic gene function analysis using the KEGG database. KOBAS software (version v2.0.12, corrected p value ≤ 0.05, kobas.cbi.pku.edu.cn) was used to verify the statistical enrichment of DEGs in KEGG pathways.
2.9. Validation of the RNAseq experiment by RT real-time PCR
Total RNA (2.5 μg) was extracted from the sample leaves and reverse transcribed as reported in (Russo et al. Citation2021). To validate the reliability of RNAseq in determining gene expression levels, real-time qRT‒PCR was performed on ten selected DEGs (Table S1). Citrus limon actin was chosen as the housekeeping gene to normalize the gene expression data. All reactions were performed in triplicate, and the fold change measurements were calculated with the 2−ΔΔCT method.
3. Results
3.1. Detection and quantification of P. tracheiphilus by real-time PCR
Since the aim of our experiment was to focus on the onset of the disease, sampling was performed 7 days after fungal inoculation. At that time, the leaves showed no symptoms in any of the considered treatments (Figure S1). To verify that the infection had occurred, the total genomic DNA was extracted from samples of both control lemon leaves (CK), leaves inoculated with P. tracheiphilus (Pt) and leaves presprayed with the biocontrol agent and then inoculated with the fungus (3CPt). Fungus DNA was detected by real-time PCR (Licciardello et al. Citation2006) either on leaf samples inoculated with P. tracheiphilus (Pt) or on those samples pretreated with bacterial suspension and inoculated with fungus (3CPt). However, the detected DNA concentrations were four times lower in 3CPt samples than in Pt leaves (Figure S2), clearly indicating that the presence of the bacterial biocontrol agent affects the replication of the fungus within the plant, at least during the asymptomatic phase of the disease. No fluorescence signals were obtained from control lemon leaves (CK) or from the PCR real-time negative controls (data not shown). Moreover, in the successive symptomatic phase, the disease index of plants pretreated with P. mediterranea strain PVCT 3C was significantly lower (p < 0.05) than that of plants inoculated exclusively with the fungal pathogen (data not shown).
3.2. Assembly and annotation of transcripts
The quality of the RNA samples (from 7 days after inoculation samples) was assessed prior to library preparation by measuring the RIN. The average RIN value was 7.3 (), suggesting that the RNA quality was suitable for downstream analyses. Raw reads were filtered out to remove reads containing adapters or low-quality reads. Further analyses were based on a total of 436 million clean reads with an average of ∼ 36 million reads (∼ 10.9 G) per sample, with average Q30 and GC percentages of 91.03% and 43.18%, respectively. The de novo assembly of the clean reads yielded 156,377 transcripts and 38,831 unigenes with N50 lengths of 2981 and 2915, respectively (), indicating that optimal transcriptome coverage was achieved. The mean mapping rate of the reads obtained by mapping back the unique filtered reads to the final assembled leaf transcriptome was 80.17% using Bowtie2 alignment software, indicating good consistency of the assembly (). To obtain a complete functional annotation of the genes, all the assembled unigenes were compared with public databases, including the National Center for Biotechnology Information (NCBI), the family of proteins (Pfam), the Cluster of Orthologous Groups of proteins (KOG/COG), SwissProt, the KEGG Orthologue (KO) and the Gene Ontology (GO) databases (Figure S3). A total of 80.89% of the unigenes were annotated in at least one of the databases used. Among them, 69.62% and 76.19% of the assembled unigenes showed identities with sequences in the Nr and Nt databases, respectively. The percentages of homologous assembled unigenes to sequences in the KO, Swiss-Prot, Pfam, GO and KOG databases were 24.37%, 49.21%, 47.17%, 47.17%, and 13.83%, respectively (Figure S3).
Table 1. General statistics of RNA quality and sequencing results.
3.3. Identification of differentially expressed genes (DEGs)
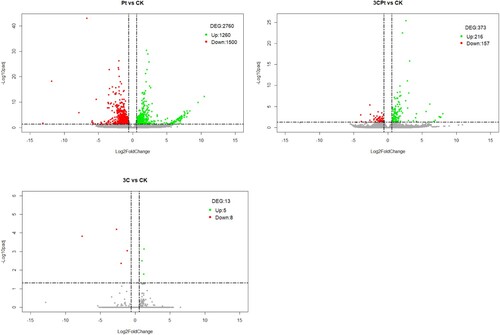
Unigenes whose expression level changed due to the treatments were identified as differentially expressed genes (DEGs) and were used to characterize the transcriptomic response of C. limon under the aforementioned experimental conditions. From the comparison of Pt vs. CK (P. tracheiphilus treated-sample set vs. control sample set), a total of 2760 differentially expressed genes were identified (1260 upregulated and 1500 downregulated) ((A)). In the case of the 3CPt vs. CK comparison (samples pretreated with the bacterial suspension and inoculated with P. tracheiphilus vs. control samples), 373 differentially expressed genes were identified (216 upregulated and 157 downregulated) ((B)). For 3C vs. CK (samples treated with the bacterial agent vs. set of control samples), 13 differentially expressed genes were identified (5 upregulated and 8 downregulated) ((C)). The analysis of these data showed that pretreatment with the bacterial biocontrol agent decisively reduced the transcriptome reprogramming induced by the fungal pathogen, at least in the asymptomatic phase of the disease (from 2760 deregulated genes to 373). In contrast, pretreatment with the biocontrol agent did not induce significant transcriptional changes, since only 13 genes were found to be differentially expressed. Most of these genes encode hypothetical proteins. Among the downregulated genes, of considerable interest is a gene that encodes an LRR receptor-like serine/threonine-protein kinase FLS2 (Cluster-6461.19974, −7.6 log2FoldChange) and an endochitinase (Cluster-6461.3363, −2.0 log2FoldChange). However, these genes did not appear among the DEGs in the 3CPt vs. CK comparison.
Figure 1. Volcano plot showing the DEGs of the (A) Pt vs. CK comparison, (B) 3CPt vs. CK comparison and (C) 3C vs. CK comparison. The upregulated genes with statistical significance are represented by the green dots, the red dots represent the downregulated genes, and the grey dots are the DEGs with -log10padj <1.3, adopting a log2FoldChange threshold of 0.58 (1.5-fold change). The X-axis corresponds to the variation in gene expression, and the Y-axis corresponds to the -log10padj.
To validate the differential gene expression obtained with RNAseq, the measurement of the expression levels of ten selected DEGs was carried out by real-time PCR. The results show a high congruence between the RNA-seq and qRT‒PCR data (coefficient of determination R2 = 0.93), indicating the reliability of RNAseq for the quantification of gene expression (Figure S4).
3.4. Functional classification of DEGs
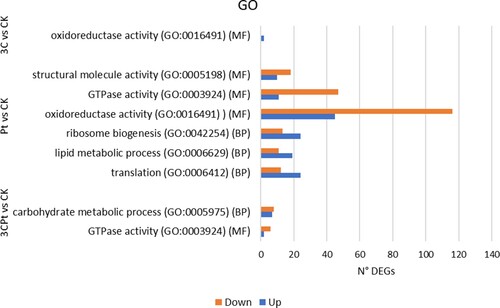
Functional enrichments of the Gene Ontology (GO) terms, of the classification Clusters of Orthologous Groups (KOG) of proteins and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were performed to identify biological processes or metabolic pathways involved in the response of the plant to the different treatments. Considering GO enrichment for the 3CPt vs. CK comparison, ‘GTPase’ (GO: 0003924) (2 upregulated and 6 downregulated) was the most enriched term in the molecular function (MF) category, while ‘Carbohydrate’ (GO: 0005975) (7 upregulated and 8 downregulated) was the most enriched term in the biological process (BP) category (). Considering GO enrichment for the Pt vs. CK comparison, ‘Translation’ (GO: 0006412) (24 upregulated and 12 downregulated), ‘Lipid metabolic process’ (GO: 0006629) (19 upregulated and 11 downregulated), and ‘Ribosome biogenesis’ (GO: 0042254) (24 upregulated and 13 downregulated) were among the most enriched terms in the biological process (BP) category, while ‘Oxidoreductase activity’ (GO: 0016491) (45 upregulated and 116 downregulated), ‘GTPase activity’ (GO: 0003924) (11 upregulated and 47 downregulated), and ‘Structural molecule activity’ (GO: 0005198) (10 upregulated and 18 downregulated) were among the most enriched terms in the molecular function (MF) category.
Figure 2. GO Gene Ontology (GO) Enrichment Analysis. The X-axis indicates the number of DEGs for each term, and the Y-axis indicates the terms represented for each comparison. BP: biological processes; MF: molecular functions.
The unigenes were aligned to the KOG database and assigned to the KOG categories (Figure S5) to categorize the possible functions. ‘Posttranslational modification, protein turnover, chaperones’ (14%) represented the largest group, followed by ‘General function prediction only’ (13.4%) and ‘Translation, ribosomal structure and biogenesis’ (11.6%). ‘Energy production and conversion’ (6.6%) and ‘Intracellular trafficking, secretion, and vesicular transport’ (6.4%) were the next most numerous categories (Figure S5).
3.5. Comprehensive analysis of the main deregulated pathways: MapMan analysis
To acquire a complete picture of the metabolic modifications that occurred in lemon leaves either in the presence of P. tracheiphilus (Pt) or during the simultaneous presence of the pathogen and P. mediterranea (3CPt), all the significant DEGs were mapped on the MapMan 3.6.0RC1 tool. reports the main ten categories related to both Pt vs. CK and 3CPt vs. CK comparisons. Overall, considering the comparison of Pt vs. CK, the analysis indicates that among the pathways specifically involved in the response to P. tracheiphilus infection, the ‘Photosynthesis’ category accounted for 1 upregulated gene and 35 downregulated genes. Another metabolic process significantly involved in the response to the pathogen was ‘Coenzyme metabolism’ with 1 upregulated and 27 downregulated genes, many of which are directly or indirectly involved in the photosynthetic process (biosynthesis of iron/sulfur proteins, biosynthesis of chlorophyll, plastoquinone biosynthesis) (data not shown), thus confirming that ‘Photosynthesis’ is the most affected metabolic pathway during fungal infection. The ‘Cellular respiration – oxidative phosphorylation’ category included 14 upregulated and 1 downregulated gene in the Pt vs. CK comparison (). The activation of oxidative phosphorylation at the mitochondrial level that is observed in plants inoculated with the fungus could precisely makeup for the energy deficiency induced by the block of the photosynthetic process. Among the DEGs of the ‘Secondary metabolism’ category (12 downregulated genes), 7 downregulated genes were in the subcategory ‘terpenoid biosynthesis’, and 5 downregulated genes were in the subcategory ‘phenolics biosynthesis’. In particular, terpenoid compounds include a wide range of substances that share the initial biosynthesis steps, such as cytokinins, abscisic acid, brassinosteroids and strigolactones. This finding suggests that the formation of these plant hormones might be deregulated in samples inoculated with the fungus. Accordingly, in the category ‘Phytohormone action’, several DEGs were found to be deregulated, indicating that the fungus induced a strong reprogramming of various signal cascades promoted by plant hormones. In contrast, none of these genes were differentially expressed in the 3CPt vs. CK comparison, indicating that pretreatment with P. mediterranea prevented, during the asymptomatic phase of the disease, the deregulation of important metabolic pathways such as photosynthesis or phytohormone action (). Furthermore, additional DEGs exclusively belonging to the Pt vs. CK comparison were within the ‘Multiprocess regulation’ (17 upregulated and 12 downregulated genes) and ‘Protein modification – phosphorylation and glycosylation’ (5 upregulated genes) categories. Instead, the ‘Cell wall organization’ (4 upregulated and 20 downregulated genes) and ‘Solute transport – primary active transport – ABC superfamily’ (11 downregulated genes) categories were deregulated in both datasets. However, the most interesting data were observed in the ‘External stimulus response to pathogen’ category, which included 6 downregulated genes exclusively in the samples inoculated with the fungus and not in those pretreated with P. mediterranea and inoculated with the fungus ().
Table 2. Number of deregulated genes belonging to various metabolic pathways and processes as a result of MapMan analysis.
Considering their importance in the transcriptome remodeling induced by fungal infection, the ‘Phytohormone action’ and ‘External stimuli-response to pathogen’ categories were analyzed in more detail. In , several genes involved in plant hormone biosynthesis and perception are listed along with their log2fold change. Abscisic-aldehyde oxidase, abscisic acid receptor PYL2 and magnesium-chelatase subunit ChlH implicated in abscisic acid biosynthesis and perception were decisively downregulated in the Pt vs. CK comparison, whereas only the abscisic-aldehyde oxidase gene was downregulated in the 3CPt vs. CK comparison. Two genes encoding enzymes involved in salicylic acid biosynthesis (phenylalanine ammonia lyase and isochorismate synthase) were also downregulated, indicating that in the Pt vs. CK comparison, both abscisic acid and salicylic acid signal transduction was affected by fungal infection. Similarly, 1-aminocyclopropane-1-carboxylate oxidase, involved in ethylene biosynthesis, was downregulated, and the ETO1-like protein 1-encoding gene, which negatively regulates ethylene production, was upregulated in the Pt vs. CK comparison, suggesting that ethylene biosynthesis is also downregulated by fungal infection (). Regarding gibberellin (GA), the GA 20 oxidase 1 gene, catalysing the late step of GA biosynthesis, is upregulated in response to fungal infection, and concordantly, several clusters involved in endogenous bioactive GA inactivation (gibberellin 2-beta-dioxygenase) are strongly downregulated, indicating that GA-induced signal transduction plays a key role in the response to mal secco disease (). Genes involved in cytokinin biosynthesis and signaling (adenylate isopentenyltransferase 5, histidine-containing phosphotransfer protein 3) were slightly induced in response to fungal infection (), as well as several genes involved in auxin biosynthesis, perception and internal distribution (). The brassinosteroid-6-oxidase-encoding gene, which synthesizes the active form of brassinosteroids, was also upregulated in the Pt vs. CK dataset. The above-described deregulation of the main phytohormone biosynthesis and signaling induced by P. tracheiphylus was not observed in the 3CPt vs. CK comparison, suggesting that pretreatment with bacterial suspension prevented it (). Among the clusters categorized within the ‘External stimuli – response to pathogens’ category, those encoding disease resistance-like protein DSC1, disease resistance RPP13-like protein 1, Leaf RUST 10 (LRK10) disease-resistance receptor-like protein kinase, and disease resistance protein SUMM2-like were downregulated specifically in the Pt vs. CK dataset (). DSC1 acts as a trigger of the hypersensitive response (HR) (Freh et al. Citation2022), RPP13-like genes play important roles in the resistance of various plant diseases (Cheng et al. Citation2018), and the disease resistance protein SUMM2-like is required for constitutive defence responses (Zhang et al. Citation2017). LRK10 is a probable receptor-like serine/threonine-protein kinase involved in abscisic acid (ABA) signaling that acts as a positive regulator of the abiotic stress response (Lim et al. Citation2015) (). Considering their roles in plant defence, these genes are to be considered of extreme interest in representing the barrier the fungus has to breakdown to overcome the plant defence during the onset of the disease.
Table 3. List of clusters in Pt vs. CK and 3CPt vs. CK comparisons.
3.6. Metabolic pathways and biological processes deregulated by fungal inoculation despite beneficial bacterium treatment
The following analysis aimed to identify clusters that were deregulated despite being subjected to BCA treatment before fungal inoculation. We counted 85 clusters in common between the Pt vs. CK and 3CPt vs. CK comparisons, and all of them were regulated concordantly and almost to the same extent (data not shown). Among them, lists key deregulated genes. In particular, abscisic-aldehyde oxidase, abscisic acid receptor PYL2, involved in abscisic acid biosynthesis and perception, and guard cell S-type anion channel SLAC1, a plasma membrane essential for stomatal closure in response to abscisic acid (Vahisalu et al. Citation2008), were strongly downregulated in both comparisons, clearly indicating that the shutdown of the abscisic acid signal cascade is a crucial factor in determining the onset of the disease. Similarly, the CTR1 gene, encoding a negative regulator of the ethylene response pathway (Kieber et al. Citation1993 ), was downregulated in both comparisons, suggesting that the transcriptomic changes in response to the fungus are directed to turn off the ethylene signaling pathway. A leucine-rich repeat receptor-like kinase MDIS1-INTERACTING RECEPTOR-LIKE KINASE 2 (MIK2), which plays a crucial role in fungal elicitor perception and in contributing to resistance against F. oxysporum in Arabidopsis (Coleman et al. Citation2021), was found to be downregulated at mal secco disease onset in both comparisons. Finally, the gene encoding plant polygalacturonase was strongly induced by fungal infection despite BCA treatment (), probably contributing to cell wall loosening and facilitating pathogen progression (Sénéchal et al. Citation2014).
Table 4. Clusters in common between the Pt vs. CK and 3CPt vs. CK comparisons.
3.7. Analysis of WRKY transcription factor family
In Figure S6, the most represented transcription factor (TF) families in terms of DEG number are reported. The results showed that a total of 133 genes encoding transcription factors were deregulated in the Pt vs. CK dataset, whereas only 18 genes were deregulated in the 3CPt vs. CK comparison. Among them, the C2H2 type, C2C2->C2C2-CO-like, MYB or MYB-related, AP2/ERF, NAC and bHLH families accounted for the highest number of deregulated genes in both comparisons. Chitin is a well-established elicitor of the plant defence response, and the gene expression eliciting activity involves the WRKY transcription factor family (Libault et al. Citation2007 ). Considering their role during pathogenesis (Russo et al. Citation2021), reports the deregulated genes belonging to this category in the Pt vs. CK dataset and compares them with the deregulated WRKY transcription factors observed in previous experiments performed on rough lemon (Russo et al. Citation2021). In that experiment, genes encoding WRKY transcription factors were among the DEGs in the fungus-inoculated vs. control sample comparison, and most of them were overexpressed (Russo et al. Citation2021). As shown in , WRKY 35, WRKY 4, WRKY 40 and WRKY 18 were also upregulated in inoculated 2Kr lemon leaves (Pt vs. CK comparison), whereas no deregulation was observed for the other WRKY genes. Moreover, those genes were not in the DEG list of the 3CPt vs. CK dataset (), suggesting that BCA pretreatment prevented their activation at this stage of the disease and caused a delay in pathogen ‘sensing’.
Table 5. Lists of WRKY transcription factors deregulated in lemon 2Kr and rough lemon (Russo et al. Citation2021).
4. Discussion
The majority of Mediterranean citrus-growing countries, with the exception of Morocco and Portugal, are included in the current geographic distribution of MSD, along with the east coast of the Black Sea (Georgia) (EPPO Global Database Citation2022). A recent study modeling the distribution of P. tracheiphilus in the Mediterranean area under future climatic scenarios supposed that climate change will likely reduce the extent of suitable sites for the spread of mal secco disease, but no range shift is expected in the Mediterranean basin that will affect the disease (Krasnov et al. Citation2022). Therefore, it is crucial to set up novel strategies to manage the disease, taking into account the need to guarantee high-quality production and environmental protection. To fill the gap in knowledge of BCA-pathogen-plant interactions at the molecular level, the aim of our work was to investigate the effect of the application of P. mediterranea suspensions on the transcriptome of lemon leaves artificially inoculated with P. tracheiphilus. RNAseq turned out to be a powerful tool revealing that even in the asymptomatic phase, the fungus induces a marked reprogramming of the transcriptome, which affects, among others, the photosynthetic process in all its phases and components. However, pretreatment with P. mediterranea limited pathogen replication and drastically reduced lemon transcriptome modifications in terms of the number of DEGs. Overall, the analysis of plant transcriptomes has shed light on plant responses to beneficial microbes in crops and in Arabidopsis as a model plant, in many cases outlining the mechanisms of action by which BCAs defend plants from pathogen attack (Feng et al. Citation2012; Duke et al. Citation2017; Kong et al. Citation2022). In particular, it has been shown that beneficial bacteria are able to generate a hostile environment for subsequent plant colonization by a virulent strain of R. solanacearum in Arabidopsis (Feng et al. Citation2012). Furthermore, the biological control agent Pseudomonas chlororaphis PA23 is capable of protecting Brassica napus (canola) from the necrotrophic fungus Sclerotinia sclerotiorum via direct antagonism (Duke et al. Citation2017), whereas Burkholderia sp. SSG suppresses pathogens by preparing and improving the defence capacity of the plant’s immune system and hormone levels (Kong et al. Citation2022). Nonpathogenic microorganisms can induce ISR (induced systemic resistance) in plants, which is able to enhance their defensive capacity against multiple plant pathogens (van Loon et al. Citation1998; Conrath et al. Citation2015). Plant defence responses may include thickening of cell walls by lignification, deposition of callose, accumulation of low-molecular-weight antimicrobial substances (e.g. phytoalexins), and synthesis of various pathogenesis-related (PR) proteins and other bacterial and fungal elicitors (Palmieri et al. Citation2022). Although leading to a similar phenotypical response, the ISR activated by beneficial bacteria requires ethylene and jasmonic acid signaling pathways, whereas pathogen-induced SAR (systemic acquired resistance) (Fu and Dong Citation2013) is mediated through salicylic acid (SA) signaling pathways (van Loon et al. Citation1998; Conrath et al. Citation2015). Based on transcriptomic data, we show that fungal infection induces the deregulation of the main plant hormone-related genes and, in particular, the downregulation of SA and ethylene signal transduction cascades, probably blocking the activation of both SAR and ISR.
To defend themselves, plants detect conserved molecules in pathogens (pathogen-associated molecular, PAMPs) via pattern recognition receptors (PRRs) to induce PAMP-triggered immunity (PTI). To cause disease, pathogens deliver effectors that suppress PTI in susceptible plants, resulting in effector-triggered susceptibility (ETS). In resistant plants, the effectors or their activities can be detected by plant R proteins, giving rise to effector-triggered immunity (ETI) (Jones and Dangl Citation2006; Chang et al. Citation2019). ETI involves an amplification of PTI defence pathways and is often associated with rapid localized cell death at the infection site, termed the hypersensitive response (HR) (Jones and Dangl Citation2006; Chang et al. Citation2019). In our work, the expression of several genes involved in the effector triggered immunity (ETI) pathway was strongly repressed. However, at the stage of the disease we looked at, BCA treatment prevented the deregulation of the genes involved in phytohormone signal cascades as well as the downregulation of the genes involved in effector-triggered immunity (ETI), suggesting that pretreatment had a powerful protective effect on the defence mechanisms of the plant. Nevertheless, genes involved in SAR or ISR were not induced in the 3CPt vs. CK comparison, suggesting that BCA drastically slowed the onset of the disease, probably by competing with fungi for space and nutrients and not by inducing plant resistance.
Plants have developed various adaptation strategies to cope with stressful conditions, and a typical example is stomatal closure limiting water loss and restricting pathogen entry into leaves. Abscisic acid is an important component of plant defence against biotic and abiotic stress and is a strong stomatal closure inducer (Puglisi et al. Citation2013; Bharath et al. Citation2021). Our results highlight the sharp downregulation of abscisic acid biosynthesis, perception and signaling, as well as the downregulation of the guard cell S-type anion channel SLAC1-encoding gene, which together account for the establishment of a pathogen-induced constraint to maintain stomatal opening, which is obviously an unfavorable status and source of further stress. This group of genes was also downregulated in the 3CPt vs. CK comparison, suggesting that they probably represent an Achille’s heel of this plant‒pathogen interaction. Similarly, the MDIS1-interacting receptor-like kinase 2-encoding gene, which is a crucial component of early immune responses to a fungal-derived elicitor (Coleman et al. Citation2021), was downregulated despite BCA treatment. Moreover, a recent study investigating the influence of P. tracheiphilus infections on the microbial communities of sour orange roots and xylem reported that bacteria belonging to the Pseudomonas and Burkholderia genera were depleted in both leaf- and root-inoculated plants, although more markedly in the rhizosphere of leaf-inoculated plants (Dimaria et al. Citation2023), thus suggesting that the decrease in beneficial microbial taxa might enhance susceptibility and favor fungal colonization in the period following the onset of the disease.
In conclusion, our results shed new light on the host‒pathogen molecular interactions during the asymptomatic phase of mal secco disease, and further studies will make it possible to plan sustainable control strategies for MSD based on the use of P. mediterranea as a biocontrol agent alone or in combination with copper-based pesticides or resistance inducers. Moreover, in this study, several genes involved in the plant response to mal secco could represent markers of the asymptomatic phase of the disease and suitable targets for genome editing applications in lemon breeding.
Supplemental Material
Download MS Word (3.1 MB)Additional information
Funding
Notes on contributors
Angelo Sicilia
Angelo Sicilia, a Ph.D. researcher in plant genetics, department of agriculture, food and environment, University of Catania. He is an expert in transcriptomic and epigenetic studies of plants of agricultural interest subjected to abiotic and biotic stress.
Vittoria Catara
Vittoria Catara, Ph.D., an associated professor in plant pathology, department of agriculture, food and environment, University of Catania. Her studies are mainly devoted to plant-associated bacteria (plant pathogens and beneficial), their interaction with the host plant and the microbiome.
Giulio Dimaria
Giulio Dimaria, a Ph.D. in Biotechnology, University of Catania, is engaged in molecular studies targeting Pseudomonas spp. as biocontrol agents against plant pathogens and on microbial multispecies interactions within the plant host.
Emanuele Scialò
Emanuele Scialò, a Ph.D. Student, department of agriculture, food and environment, University of Catania. He is actually involved in transcriptomic analyses of the Citrus plant response to environmental challenges.
Marcella Russo
Marcella Russo, a Ph.D., Research and Services Agrobiotech Soc. Coop., Catania, Italy. She is a researcher in plant pathology responsible for the diagnostic laboratory.
Alessandra Gentile
Alessandra Gentile, a Ph.D., department of agriculture, food and environment, University of Catania. Her main interest is the genetic improvement of fruit trees by traditional and innovative methods (considerable expertise through biotechnological approaches).
Angela Roberta Lo Piero
Angela Roberta Lo Piero, a Ph.D., professor in plant genetics, department of agriculture, food and environment, University of Catania. She is an expert in the identification of transcriptome and epigenome modifications induced by both biotic and abiotic stresses, in particular, those related to climate changes.
References
- Aiello D, Leonardi GR, Di Pietro C, Vitale A, Polizzi G. 2022. A new strategy to improve management of citrus mal secco disease using bioformulates based on Bacillus amyloliquefaciens strains. Plants. 11:446. doi: 10.3390/plants11030446.
- Batuman O, Ritenour M, Vicent A, Li H, Hyun J, Catara V, Ma H, Cano LM. 2020. Diseases caused by fungi and oomycetes. In: Talón M, Caruso M, Gmitter FG Jr, editors. The genus Citrus. Amsterdam: Elsevier; p. 349–369.
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 57:289–300. doi:10.1111/j.2517-6161.1995.tb02031.x.
- Bharath P, Gahir S, Raghavendra AS. 2021. Abscisic acid-induced stomatal closure: an important component of plant defense against abiotic and biotic stress. Front Plant Sci. 12:615114. doi: 10.3389/fpls.2021.615114.
- Catalano C, Di Guardo M, Distefano G, Caruso M, Nicolosi E, Deng Z, Gentile A, La Malfa SG. 2021. Biotechnological approaches for genetic improvement of lemon (Citrus limon (L.) Burm. f.) against mal secco disease. Plants. 10:1002. doi:10.3390/plants10051002.
- Catara A, Catara V. 2019. Il “mal secco” degli agrumi: da un secolo in Sicilia. Memorie e Rendiconti USPI Associato all'Unione Stampa Periodica Italiana; Giovanni Battista RM. 3:33–58.
- Catara V. 2007. Pseudomonas corrugata: plant pathogen and/or biological resource? Mol Plant Pathol. 8:233. doi: 10.1111/j.1364-3703.2007.00391.x.
- Catara V, Sutra L, Morineau A, Achouak W, Christen R, Gardan L. 2002. Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea sp. nov. Int J Syst Evol Microbiol. 52:1749–1758. doi:10.1099/00207713-52-5-1749.
- Chang M, Clinton M, Liu F, Fu ZQ. 2019. NAD+ cleavage: TIR domain-containing resistance proteins in action. Trends Plant Sci. 24:1069. doi: 10.1016/j.tplants.2019.10.005.
- Cheng J, Fan H, Li L, Hu B, Liu H, Li Z. 2018. Genome-wide identification and expression analyses of RPP13-like genes in barley. Biochip J. 12:102. doi: 10.1007/s13206-017-2203-y.
- Coco V, Grimaldi V, Catara V, Licciardello G, Cirvilleri G, Grasso S, Catara A. 2004. Inhibition of Phoma tracheiphila by Pseudomonads in citrus seedlings. Proc Int Soc Citriculture. 2:729–732.
- Coleman AD, Maroschek J, Raasch L, Takken FLW, Ranf S, Hückelhoven R. 2021. The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytol. 229(6):3453–3466. doi: 10.1111/nph.17122.
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR. 2015. Priming for enhanced defense. Annu Rev Phytopathol. 53:97. doi:10.1146/annurev-phyto-080614-120132.
- Corke ATK. 1974. Biological control of plant pathogens. By Baker KF and James Cook R. San Francisco: Freeman; p. 433.
- Deng H, Li XF, Cheng WD, Zhu YG. 2009. Resistance and resilience of Cu-polluted soil after Cu perturbation, tested by a wide range of soil microbial parameters. FEMS Microbiol Ecol. 70(2):293–304. doi: 10.1111/j.1574-6941.2009.00741.x.
- Dimaria G, Mosca A, Anzalone A, Paradiso G, Nicotra D, Privitera GF, Pulvirenti A, Catara V. 2023. Sour orange microbiome is affected by infections of Plenodomus tracheiphilus causal agent of citrus mal secco disease. Agronomy. 13(3):654. doi: 10.3390/agronomy13030654.
- Duke KA, Becker MG, Girard IJ, Millar JL, Fernando D, Belmonte WG, de Kievit TR. 2017. The biocontrol agent Pseudomonas chlororaphis PA23 primes Brassica napus defenses through distinct gene networks. BMC Genom. 18(1):467. doi: 10.1186/s12864-017-3848-6.
- EPPO Global Database. 2022. [accessed 2022 Oct 17]. https://gd.eppo.int/taxon/DEUTTR.
- Feng DX, Tasset C, Hanemian M, Barlet X, Hu J, Trémousaygue D, Deslandes L, Marco Y. 2012. Biological control of bacterial wilt in Arabidopsis thaliana involves abscissic acid signalling. New Phytol. 194:1035–1045. doi: 10.1111/j.1469-8137.2012.04113.x.
- Freh M, Gao J, Petersen M, Panstruga R. 2022. Plant autoimmunity—fresh insights into an old phenomenon. Plant Physiol. 188:1419. doi: 10.1093/plphys/kiab590.
- Fu ZQ, Dong X. 2013. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 64(1):839–863. doi: 10.1146/annurev-arplant-042811-105606.
- Gentile A, La Malfa SG. 2019. New breeding techniques for sustainable agriculture. In: Book: innovations in sustainable agriculture; p. 411–437. doi: 10.1007/978-3-030-23169-9_13.
- Ghadamgahi F, Tarighi S, Taheri P, Saripella GV, Anzalone A, Kalyandurg PB, Catara V, Ortiz R, Vetukuri RR. 2022. Plant growth-promoting activity of Pseudomonas aeruginosa FG106 and its ability to act as a biocontrol agent against potato, tomato and taro pathogens. Biology. 11(1):140. doi: 10.3390/biology11010140.
- Jones JD, Dangl JL. 2006. The plant immune system. Nature. 444(7117):323–329. doi: 10.1038/nature05286.
- Kelbessa BG, Dubey M, Catara V, Ghadamgahi F, Ortiz R, Vetukuri RR. 2023. Potential of plant growth-promoting rhizobacteria to improve crop productivity and adaptation to a changing climate; p. 1–14. doi: 10.1079/cabireviews202318001.
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. 1993. CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 72(3):427–441. doi:10.1016/0092-8674(93)90119-B.
- Kong P, Li X, Gouker F, Hong C. 2022. cDNA transcriptome of Arabidopsis reveals various defense priming induced by a broad-spectrum biocontrol agent Burkholderia sp. SSG. Int J Mol Sci. 23(6):3151. doi: 10.3390/ijms23063151.
- Krasnov H, Ezra D, Bahri BA, Cacciola SO, Meparishvili G, Migheli Q, Blanket L. 2022. Potential distribution of the citrus Mal Secco disease in the mediterranean basin under current and future climate conditions. Plant Pathol. 72:765–773. doi:10.1111/ppa.13692.
- Lahlali R, Ezrari S, Radouane N, Kenfaoui J, Esmaeel Q, EL Hamss H, Belabess Z, Barka EA. 2022. Biological control of plant pathogens: a global perspective. Microorganisms. 10(3):596. doi: 10.3390/microorganisms10030596.
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. 2007. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact. 20(8):900–911. doi:10.1094/MPMI-20-8-0900.
- Licciardello G, Grasso FM, Bella P, Cirvilleri G, Grimaldi V, Catara V. 2006. Identification and detection of Phoma tracheiphila, causal agent of citrus mal secco disease, by realtime polymerase chain reaction. Plant Dis. 90(12):1523–1530. doi: 10.1094/PD-90-1523.
- Lim CW, Yang SH, Shin KH, Lee SC, Kim SH. 2015. Probable receptor-like serine/threonine-protein kinase involved in abscisic acid (ABA) signaling. Plant Cell Rep. 34(3):447–455. doi: 10.1007/s00299-014-1724-2.
- Migheli Q, Cacciola SO, Balmas V, Pane A, Ezra D, di San Lio GM. 2009. Mal secco disease caused by Phoma tracheiphila: a potential threat to lemon production worldwide. Plant Dis. 93(9):852–867. doi: 10.1094/PDIS-93-9-0852.
- Nigro F, Ippolito A, Salerno MG. 2011. Mal secco disease of citrus: a journey through a century of research. J Plant Pathol. 93:523.
- Oliveri C, Modica G, Bella P, Continella A, Catara V. 2022. Preliminary evaluation of a zinc-copper-citric acid biocomplex for the control of Plenodomus tracheiphilus causal agent of citrus mal secco disease. Acta Hortic. 1354:231.
- Palmieri D, Ianiri G, Del Grosso C, Barone G, De Curtis F, Castoria R, Lima G. 2022. Advances and perspectives in the use of biocontrol agents against fungal plant diseases. Horticulturae. 8(7):577. doi:10.3390/horticulturae8070577.
- Pérez-Tornero O, Córdoba F, Moreno M, Yuste L, Porras I. 2012. Classic methods and biotechnical tools in lemon breeding: preliminary results. Acta Hortic. 928:259–263. doi: 10.17660/ActaHortic.2012.928.32.
- Poles L, Licciardello C, Distefano G, Nicolosi E, Gentile A, La Malfa S. 2020. Recent advances of in vitro culture for the application of new breeding techniques in citrus. Plants. 9(8):938. doi: 10.3390/plants9080938.
- Puglisi I, Lo Cicero L, Lo Piero AR. 2013. The glutathione S-transferase gene superfamily: an in silico approach to study the post translational regulation. Biodegradation. 24(4):471–485. doi: 10.1007/s10532-012-9604-3.
- Raymaekers K, Ponet L, Holtappels D, Berckmans B, Cammue BPA. 2020. Screening for novel biocontrol agents applicable in plant disease management – a review. Biol Control. 144:104240. doi: 10.1016/j.biocontrol.2020.104240.
- Russo M, Grasso FM, Bella P, Licciardello G, Catara A, Catara V. 2011. Molecular diagnostic tools for the detection and characterization of Phoma tracheiphila.». Acta Hortic. 892:207–214. doi: 10.17660/ActaHortic.2011.892.24.
- Russo R, Caruso M, Arlotta C, Lo Piero AR, Nicolosi E, Di Silvestro S. 2020. Identification of field tolerance and resistance to mal secco disease in a citrus germplasm collection in Sicily. Agronomy. 10(11):1806. doi: 10.3390/agronomy10111806.
- Russo R, Sicilia A, Caruso M, Arlotta C, Di Silvestro S, Gmitter FG Jr, Nicolosi E, Lo Piero AR. 2021. De novo transcriptome sequencing of rough lemon leaves (Citrus jambhiri lush.) in response to Plenodomus tracheiphilus infection. Int J Mol Sci. 22(2):882. doi: 10.3390/ijms22020882.
- Sénéchal F, Wattier C, Rustérucci C, Pelloux J. 2014. Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot. 65(18):5125–5160. doi: 10.1093/jxb/eru272.
- Sicilia A, Russo R, Caruso M, Arlotta C, Di Silvestro S, Gmitter FG Jr, Gentile A, Nicolosi E, Lo Piero AR. 2022. Transcriptome analysis of Plenodomus tracheiphilus infecting rough lemon (Citrus jambhiri Lush.) indicates a multifaceted strategy during host pathogenesis. Biology 11:761. doi:10.3390/biology11050761
- Sicilia A, Santoro DF, Testa G, Cosentino SL, Lo Piero AR. 2020. Transcriptional response of giant reed (Arundo donax L.) low ecotype to long-term salt stress by unigene-based RNAseq. Phytochemistry. 177:112436. doi: 10.1016/j.phytochem.2020.112436.
- Sicilia A, Testa G, Santoro DF, Cosentino SL, Lo Piero AR. 2019. RNASeq analysis of giant cane reveals the leaf transcriptome dynamics under long-term salt stress. BMC Plant Biol. 19(1):355. doi: 10.1186/s12870-019-1964-y.
- Springer NM. 2010. Isolation of plant DNA for PCR and genotyping using organic extraction and CTAB. Cold Spring Harb Protoc. 2010(11). doi: 10.1101/pdb.prot5515.
- Strano CP, Bella P, Licciardello G, Caruso A, Catara V. 2017. Role of secondary metabolites in the biocontrol activity of Pseudomonas corrugata and Pseudomonas mediterranea. Eur J Plant Pathol. 149(1):103–115. doi: 10.1007/s10658-017-1169-x.
- Talon M, Gmitter FG. 2008. Citrus genomics. Int J Plant Genom. article ID:528361. doi:10.1155/2008/528361.
- Tamm L, Thuerig B, Apostolov S, Blogg H, Borgo E, Corneo PE, Fittje S, de Palma M, Donko A, Experton C, et al. 2022. Use of copper-based fungicides in organic agriculture in twelve European countries. Agronomy. 12(3):673. doi: 10.3390/agronomy12030673.
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al. 2008. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 452(7186):487–491. doi: 10.1038/nature06608.
- van Loon LC, Bakker PAHM, Pieterse CMJ. 1998. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 36(1):453–483. doi: 10.1146/annurev.phyto.36.1.453.
- Walsh UF, Morrissey JP, O’Gara F. 2001. Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr Opin Biotechnol. 12(3):289–295. doi: 10.1016/S0958-1669(00)00212-3.
- Zhang Z, Liu Y, Huang H, Gao M, Wu D, Kong Q, Zhang Y. 2017. The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep. 18:292.
