ABSTRACT
A number of studies have attempted to elucidate the binding mechanism between tumor necrosis factor (TNF) and clinically relevant antagonists. None of these studies, however, have been conducted as close as possible to physiologic conditions, and so the relationship between the size distribution of TNF-antagonist complexes and the antagonists' biological activity or adverse effects remains elusive. Here, we characterized the binding stoichiometry and sizes of soluble TNF-antagonist complexes for adalimumab, infliximab, and etanercept that were formed in human serum and in phosphate-buffered saline (PBS). Fluorescence-detected sedimentation velocity analytical ultracentrifugation analyses revealed that adalimumab and infliximab formed a range of complexes with TNF, with the major complexes consisting of 3 molcules of the respective antagonist and one or 2 molcules of TNF. Considerably greater amounts of high-molecular-weight complexes were detected for infliximab in human serum. The emergence of peaks with higher sedimentation coefficients than the adalimumab monomer as a function of added human serum albumin (HSA) concentration in PBS suggested weak reversible interactions between HSA and immunoglobulins. Etanerept exclusively formed 1:1 complexes with TNF in PBS, and a small amount of complexes with higher stoichiometry was detected in human serum. Consistent with these biophysical characterizations, a reporter assay showed that adalimumab and infliximab, but not etanercept, exerted FcγRIIa- and FcγRIIIa-mediated cell signaling in the presence of TNF and that infliximab exhibited higher potency than adalimumab. This study shows that assessing distribution profiles in serum will contribute to a more comprehensive understanding of the in vivo behavior of therapeutic proteins.
Introduction
Since tumor necrosis factor (TNF) was established as one of the key mediators in the pathogenesis of several immune-mediated inflammatory diseases, the therapeutic effects of various TNF antagonists, including adalimumab (Ada), infliximab (Inf), and etanercept (Eta), have been demonstrated. Adalimumab is a human IgG1 monoclonal antibody (mAb),Citation1,2 while infliximab is a chimeric mAb consisting of the TNF-binding site derived from the murine mAb A2 linked to the constant region of human IgG1.Citation3,4 Etanercept is a recombinant fusion protein composed of a human IgG1 Fc fragment fused to the extracellular ligand-binding portion of the human TNF receptor (TNFR) p75.Citation5
Adalimumab, infliximab, and etanercept are effective in treating rheumatoid arthritis, psoriatic arthritis, ankylosing spondylitis, and psoriasis.Citation2,4-8 In contrast to etanercept, adalimumab and infliximab have demonstrated therapeutic effects in treating Crohn's disease and the clinical efficacy of the 2 has been estimated to be similar.Citation9-11
Common side effects associated with TNF antagonists are usually mild and well tolerated, and the incidence and severity varies slightly between adalimumab, infliximab, and etanercept.Citation12 Immunogenicity is another concern for TNF antagonists, and this can result in diminished efficacy and increased side effects. As with other pharmaceutical formulations, the therapeutic agents studied here can interfere with the immune system and might trigger immune responses leading to the formation of anti-drug antibodies (ADA). The incidence of immunogenicity in patients treated with TNF antagonist varies greatly among different studies. In psoriasis patients receiving infliximab or adalimumab, ADA have been observed at rates of 5.4–43.6% and 8.8–44.8%,Citation13 whereas for the full range of Ada and Inf-treatable diseases the occurrence was 7–68%Citation14,15 and 0.04–87%,Citation16 respectively. In the case of etanercept, low levels of ADA were consistently measured and it was confirmed that they have no effect on the drug's efficacy and safety, whereas antibodies against infliximab and adalimumab were associated with decreased clinical response.Citation13
Several hypotheses have been posited in an attempt to explain reported differences in the effectiveness of the 3 TNF antagonists in the treatment of various diseases. In addition to possible differences in pharmacokinetics and tissue distribution, the most likely explanation is related to differences in their ability to form complexes with TNF.
Each agent has been shown to have a strong intrinsic binding affinity for soluble TNF. The affinities, as determined by a surface plasmon resonance (SPR), were 30.4 pM for adalimumab, 27.3 pM for infliximab, and 11.8 pM for etanercept.Citation17 In another study, a picomolar binding affinity of adalimumab to recombinant human TNF was confirmed using bio-layer interferometry (BLI) with an estimated dissociation equilibrium constant (Kd) value of ∼4.6 pM.Citation18 Recently, Ogura et al. reported the Kds for etanercept, infliximab, and adalimumab as 5.66 pM, 88.6 pM, and 277 pM, respectively, as measured by SPR.Citation19
Size distributions and stoichiometry of complexes formed between TNF and 3 different antagonists have been characterized in vitro. A number of complexes with molecular weights (MWs) in the 600–5,800 kDa range formed between adalimumab and TNF. However, these larger complexes were found to be transient, and upon overnight incubation, a single stable complex with an MW of ∼598 kDa, consisting of 3 adalimumab and 3 TNF molecules, was identified.Citation20 Similarly, in another study it was shown that adalimumab formed a variety of complexes with TNF with MWs of up to 4,000 kDa.Citation21 In the case of infliximab, the MWs of the complexes were as high as 14,000 kDa.Citation21 It has been suggested that the most thermodynamically stable complex is composed of 6 molcules of infliximab and 3 molcules of TNF.Citation22 When infliximab was present in molar excess over TNF, complex formation between 3 molcules of infliximab and one TNF trimer was detected.Citation23 In contrast to the mAbs, etanercept did not form large complexes; only 1:1 complexes with an apparent MW of 180 kDa were identified in the presence of excess TNF.Citation21-23 When etanercept was present in excess, complex formation between 2 etanercept molecules and a single trimeric TNF (forming a 300 kDa complex) was observed.Citation21 In another study, however, etanercept bound to TNF in a 1:1 stoichiometry, even when present in molar excess.Citation23
The above-mentioned studies have clearly revealed that different TNF antagonists form various types of complexes. These findings, however, cannot be easily extrapolated to the in vivo environment to explain differences in the clinical efficacy of different TNF antagonists. Size-exclusion chromatography (SEC) coupled with light scattering (LS) or refractive index (RI) detectors and dynamic light scattering (DLS) techniques that were used in these studies require relatively simple solutions where only the molecule of interest and its interaction partner are present. Additionally, analysis is often restricted by the small number of amenable solvents, which are usually limited to general solvents such as phosphate buffers. Nevertheless, Demeule et al. showed that different complexes between a recombinant humanized mAb and its antigen can form in serum and phosphate-buffered saline (PBS).Citation24 Due to technical limitations, characterization of TNF-antagonists complexes was only performed in the micromolar concertation range.
The present study aimed to reveal binding characteristics of adalimumab, infliximab, and etanercept to recombinant human TNF under near-physiologic concentrations and solution environment conditions. The sedimentation velocity analytical ultracentrifugation (SV AUC) with absorbance (UV) detection conducted at the micromolar range showed that infliximab formed the largest complexes, followed by adalimumab, and the smallest complexes were detected with etanercept, which is consistent with previously reported findings. The next target drug concentration (25 nM) was chosen based on actual serum concentrations measured in patients.Citation2,4,5 Complexes that formed in the presence of TNF at 3 concentrations from 2.5 to 25 nM (assuming TNF is in its trimer form) were analyzed using a fluorescence detection system (FDS) coupled with SV AUC. AUC has become a widely accepted method for accurate determination of size distributions of macromolecules in solution.Citation25-28 Compared with previously used SEC and DLS methods, AUC is capable of providing higher resolution, is applicable for a virtually unlimited variety of solvent compositions, and quantification is not affected by the presence of large aggregates.Citation29-32 When coupled with the recently developed FDS, AUC has the additional advantage of allowing measurements to be performed in nanomolar and picomolar concentration ranges.Citation33-36 SV measurements using current commercially available FDS require chemical labeling of the target macromolecule with fluorescent labels with excitation maxima at 488 nm and emission at 505–565 nm. From several suitable fluorescent dyes, we chose Alexa Fluor 488 owing to its high labeling efficiency.Citation37 To confirm the integrity and TNF-binding capacity of Alexa Fluor 488-labeled antagonists, SV experiments were first performed in PBS where ideal sedimentation behavior is usually observed. Additionally, using the unprecedented ability of FDS to detect sedimentation in highly non-ideal, crowded solution environments, SV experiments were conducted in human serum.Citation34,38 To assign the peaks yielded by conventional continuous c(s) distribution modeling with SEDFIT,Citation39 SV data were further analyzed using the hybrid local continuous distribution and global discrete species model of SEDPHAT.Citation40 The stoichiometries of the derived complexes were corroborated by native mass spectrometry (MS) measurements. A dependence of sedimentation coefficient distribution on the TNF mixing ratio was observed. To explain this, a theory was proposed whose trends were confirmed by simulation data generated using adalimumab-Fab-TNF dissociation constant of 11.6 nM as estimated by isothermal titration calorimetry (ITC). Based on the differences in complex formation revealed by AUC and the different abilities to activate FcγRIIa and FcγRIIIa demonstrated using a reporter cell assay, a possible mechanism responsible for the differences in biological activity of various TNF antagonists is discussed.
Results
Interaction analysis in PBS
Recombinant human TNF purified from yeast or E. coli was shown to be present in its trimeric form in previous crystallographicCitation41 and AUC studies.Citation42 To confirm the oligomeric state of human TNF produced recombinantly in the baculovirus expression system used in this study, UV-SV AUC was performed using samples at a concentration of 2 μM. Sedimentation coefficient distributions revealed a main peak, amounting to more than 96% of the total signal intensity, with a sedimentation coefficient s20,w of 3.9 S and an estimated MW of 52 kDa, consistent with the calculated molecular weight of a trimer (Fig. S1A). Similar results were obtained in the control experiments performed with recombinant human TNF prepared using an E. coli expression system (Fig. S1B). Furthermore, FDS-SV with fluorescently labeled baculovirus-expressed TNF confirmed that it exists primarily as a stable, soluble trimer over a 2.5–25 nM concentration range in both PBS (Fig. S1C) and human serum (Fig. S1D). Accordingly, we used this concentration for trimeric TNF throughout the remainder of the manuscript.
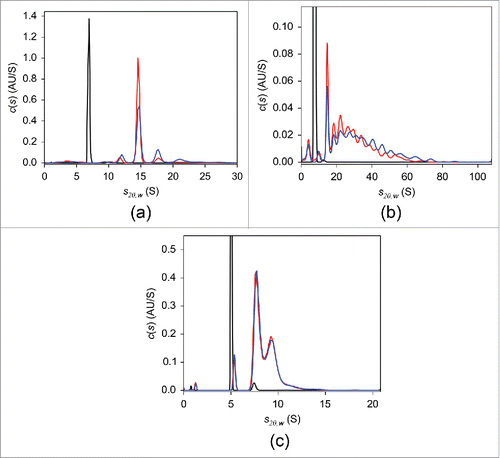
As mentioned above, a preparatory step to fluorescently label the TNF antagonists is required for the FDS-SV experiments. Prior to conjugation of fluorescent probe, the ability of native, non-labeled adalimumab, infliximab, and etanercept to form complexes with TNF was evaluated using UV-SV (). At a concertation of 2 μM, in the absence of TNF, all antagonists revealed single-peaked c(s) distributions. The peak sedimentation coefficient s20,w and estimated MW were 6.9 S and 144 kDa for adalimumab and 7.4 S and 141 kDa for infliximab, consistent with the monomeric form of the respective antibody. In the case of etanercept, a major peak was at 5.0 S with an estimated MW of 119 kDa, which is less than the 150 kDa expected for monomeric etanercept.Citation5 The frictional ratio value derived from the analysis was 2.0, in contrast to 1.4 for adalimumab and infliximab, indicating that etanercept has a more asymmetric shape. It is thought that the main underlying reason for the expanded conformation of etanercept is heavy glycosylation in the Fc region.Citation43 We concluded that the elongated structure of etanercept affected its sedimentation behavior and led to a decreased apparent sedimentation coefficient, which in turn resulted in an underestimated MW. Neither experimental nor structural data allowing for hydrodynamic calculation of sedimentation coefficient are available, which precludes unambiguous identification of the 5.0 S peak. We tentatively concluded that this peak corresponds to the monomer. When TNF at equimolar concentrations was added to antagonist solutions, peaks with sedimentation coefficients larger than those of the monomeric antagonists corresponding to complexes were observed. The weight-average sedimentation coefficients indicated that the largest complexes with TNF were formed by infliximab, whereas considerably smaller complexes were detected in adalimumab and etanercept mixtures (Table S1). Similar results were obtained using DLS, where the average hydrodynamic radius of each antagonist in solution increased with increasing molar ratios of added TNF, indicating formation of antagonist-TNF complexes. The most prominent increase was detected for infliximab, followed by adalimumab and etanercept (Fig. S2). In agreement with the previously published SEC findings,Citation20 our UV-SV results indicated that overnight incubation of adalimumab-TNF mixture at 37°C led to the formation of a single major complex with s20,w of 14.6 S (). Even though a slight decrease in populations of complexes with large sedimentation coefficients was observed following overnight incubation, in contrast to adalimumab, infliximab-TNF complexes exhibited broad continuous sedimentation coefficient distributions (). No substantial changes in the c(s) distribution of etanercept-TNF complexes were detected in response to overnight incubation ().
Figure 1. UV-SV with 2 μM adalimumab (a), infliximab (b), and etanercept (c). The c(s) distributions in the absence of TNF (black) and of the 1:1 molar mixtures of the respective antagonist:TNF incubated at 20°C for 2 hours (blue) or at 37°C overnight (red) are shown.
Next, we labeled adalimumab, infliximab, and etanercept with Alexa Fluor 488 fluorescent dye. Previous studies have clearly demonstrated that the FDS is capable of providing accurate data that are consistent with measurements conducted at micromolar concentrations using conventional absorption optics.Citation44 Therefore, the characterization of labeled TNF antagonists was directly performed at nanomolar concentrations using FDS.
To evaluate whether the covalently attached fluorescent probe affected protein structure or stability, and to confirm that the fluorescently labeled antibodies retained TNF-binding capacity, adalimumab, infliximab, and etanercept at 25 nM were subjected to FDS-SV experiments in PBS. To prevent non-specific adsorption of protein to the surfaces, such as centerpiece walls and windows, 0.1 mg/mL lysozyme was added to the solutions.
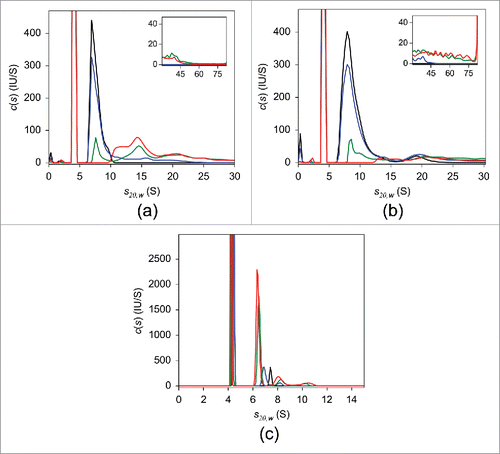
The results of SV analysis of adalimumab are shown in . Similar to the UV-SV findings, in the absence of TNF, a single peak with sedimentation coefficient s20,w of 6.8 S and estimated MW of ∼150 kDa, consistent with monomeric adalimumab, was observed in the c(s) distribution. Mixing adalimumab and TNF at a molar ratio of 10:1 induced an ∼10% reduction in uncomplexed adalimumab. When the mixing ratio was 2:1 adalimumab to TNF, the level of uncomplexed adalimumab was less than 50%. In the presence of an equimolar concentration of TNF, no free monomeric adalimumab was observed and the major adalimumab:TNF complexes were detected at 8.2 S, 11.9 S, 14.4 S, and 17.4 S (). In equimolar mixtures of infliximab and TNF, the major complex peak was at 15.2 S (). A nearly exclusive 7.0 S peak appeared in the case of etanercept (). The consistency between these and UV-SV results indicates that there was no considerable effect of fluorescent labeling on the interaction of antagonists with TNF. Detailed FDS-SV results obtained for infliximab and etanercept are described further below.
Figure 2. FDS-SV with 25 nM adalimumab (a), infliximab (b), and etanercept (c) in the presence of varying concentrations of TNF in PBS. The c(s) distributions in the absence of TNF (black) and of the 10:1 (blue), 2:1 (green), and 1:1 (red) molar mixtures of the respective antagonist:TNF are shown.
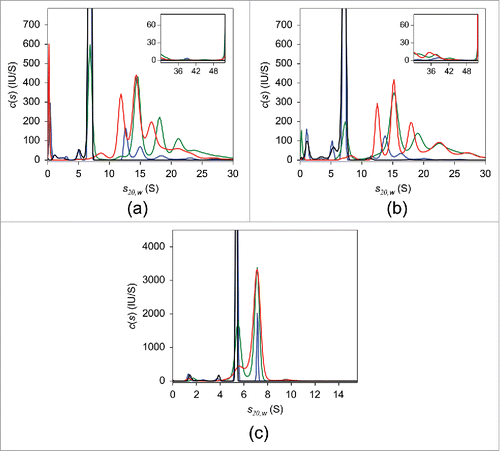
Table 1. TNF-antagonist complexes detected using native MS and AUC FDS-SV.
To compare the sizes of various complexes detected in the c(s) distributions of mixtures of the different antagonists and TNF, the weight-average sedimentation coefficients were calculated for each experimental condition (). The resulting values indicated that infliximab was capable of forming the largest soluble complexes with TNF, whereas the smallest complexes were detected in etanercept-TNF mixtures, in agreement with DLS results.
Table 2. The weight-average sedimentation coefficients calculated for each experimental condition.
To evaluate the stoichiometric ratio of adalimumab to TNF in the 4 major complexes, SV data were further analyzed in SEDPHAT using the hybrid local continuous distribution and global discrete species model. To define parameter values for discrete species, each of the 4 above-mentioned sedimentation coefficients was combined with calculated MWs, using 148 kDa for adalimumab and 55 kDa for TNF, according to values specified in the product monographCitation2 and measured by MS, respectively. The continuous distribution segment range was set to 20–50 S. Several models comprising different reasonable combinations of sedimentation coefficients and MWs reflecting different stoichiometries of adalimumab-TNF complexes were tested (Table S2). Assuming that the minimum root mean-square deviation (rmsd) indicates the model that best fits the data, the major complexes are likely to correspond to (Ada)1(TNF)1, (Ada)2(TNF)1, (Ada)3(TNF)1, and (Ada)3(TNF)2. Three other acceptable models, for which the rmsd values do not exceed the critical value, are shown in Table S2.
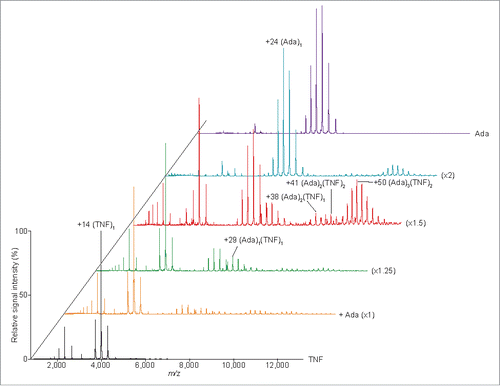
Peak assignments based on MWs derived from SV data remain ambiguous because a relatively low degree of accuracy is anticipated for continuous distributions, although for single-peak distributions the estimations can be expected to fall within 10% of the true MW value.Citation40,45 Conversely, MS under non-denaturing conditions has been shown to be a powerful approach for estimating the stoichiometry of non-covalently bound protein complexes.Citation46-49 Therefore, to verify our AUC results, the stoichiometry of adalimumab-TNF complexes was evaluated using MS under non-denaturing conditions (). (Ada)1(TNF)1, (Ada)2(TNF)1, (Ada)2(TNF)2, and (Ada)3(TNF)2 complexes were detected in experiments with 2–4 µM adalimumab and 2 µM TNF, corroborating the peak assignment determined by SV analysis ().
Figure 3. Mass spectra of the TNF-adalimumab complexes. Mass spectra of the mixtures containing trimeric TNF:adalimumab at molar ratios of 1:0 (black), 1:1 (orange), 1:1.25 (green), 1:1.5 (red), 1:2 (cyan), and 0:1.5 (purple) are shown.
In the 25 nM equimolar mixture of adalimumab and TNF, the area under the 11.9 S-peak was larger compared with the 2:1 adalimumab:TNF mixture. In addition, an 8.2 S-peak only appeared in the equimolar mixture and was not detected when adalimumab and TNF were mixed at a 2:1 molar ratio. Similar results were obtained in experiments performed with 50 nM (Fig. S3A) and 100 nM of adalimumab (Fig. S3B). When adalimumab and TNF were mixed at molar ratios of 2:1, 1:1, and 1:2, major peaks were detected at 11.9 S and 14.4 S. Again, a smaller peak at 8.4 S was detected only in the presence of equimolar or excess amounts of TNF. In the c(s) distributions of the 2:1 molar mixtures of adalimumab:TNF, the largest major complex was detected at 18.2 S, whereas with ratios of 1:1 and 1:2 molar this peak was absent, and the 17.0 S-peak was observed instead. As discussed above, the 17.0 S peak is likely to represent (Ada)3(TNF)2, whereas the 18.2 S peak corresponds to a larger complex. These results suggest that a molar excess of TNF over adalimumab results in the formation of smaller complexes.
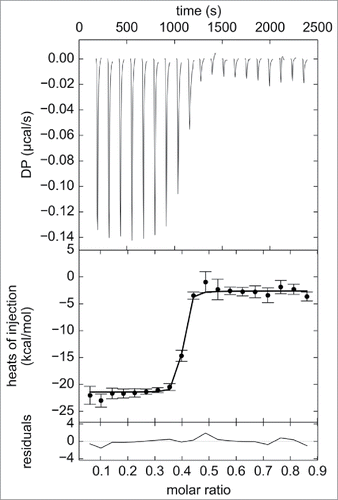
To verify that the observed changes in complex sizes were due to changes in the relative concentrations of adalimumab and TNF, numerical simulations using the Solver add-in for Microsoft Excel were performed.Citation50 It was assumed that one adalimumab (antibody, Ab) molecule is capable of binding up to 2 TNF (antigen, Ag) molecules, and one Ag molecule is capable of binding up to 3 Ab molecules in a sequential manner. The maximum number of Ab molecules participating in the interaction was limited to 3, which is supported by the MS results obtained in this study. The dissociation constants of the 9 possible AbAg complexes AbAg, AbAg2, Ab2Ag, Ab2Ag2, Ab2Ag3, Ab3Ag, Ab3Ag2, Ag3Ag3, and Ab3Ag4 were expressed as functions of molar fractions of each of the components in the solution. Together with the total Ab and Ag concentration constraints, the derived equations were used to determine the best-fit populations of each AbAg complex in the mixtures containing 25 nM Ab and 2.5–100 nM Ag by nonlinear regression analysis. All 9 dissociation constant values were assumed to be equal to that derived from the ITC analysis of direct titration of TNF into an adalimumab-Fab solution. The raw heat signals were integrated to generate a plot of ΔH kcal/mol of injected TNF versus the molar ratio of TNF/Fab, as displayed in the middle panel of . Non-linear regression analysis of the curves yielded the dissociation constant of 11.6 ± 1.7 nM, as determined from triplicate measurements. ITC experiments were also performed using full-length adalimumab, and a somewhat smaller value of 2.2 nM was obtained (Fig. S4).
Figure 4. ITC analysis of direct titration of 39.8 μM trimeric TNF into 9.8 μM adalimumab-Fab. Raw heat changes (top panel), normalized heat changes with best-fit values (solid line) (middle panel), and residuals of the fit (bottom panel) are shown. Non-linear regression analysis of the curves using A + B + B + B <-> AB + B + B <-> ABB + B <-> ABBB binding model yielded the Kd of 11.6 ± 1.7 nM and ΔH values of 11.6 ± 1.7 nM and -6.4 ± 0.1 kcal/mol, respectively, as determined from triplicate measurements. Representative profile from triplicate measurements is shown.
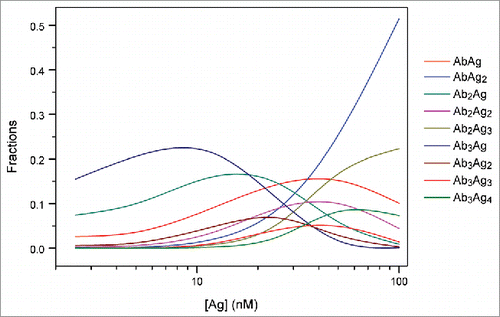
With the exception of the Ab3Ag4 complex, the simulations predicted dependence similar to that observed in AUC, indicating that at equimolar and higher concentrations of TNF, populations of complexes containing 3 molcules of adalimumab decreased, accompanied by an increase in the amount of complexes composed of one or 2 adalimumab molecules ().
Figure 5. Simulation analysis of the populations of each AbAg complex in an Ab-Ag mixture. Simulations were performed for the mixtures containing 25 nM Ab and 2.5–100 nM Ag using Kd of 11.6 ± 1.7 nM derived from the ITC analysis of direct titration of TNF into an adalimumab-Fab solution.
Next, we performed FDS-SV with 25 nM of infliximab (). In the absence of TNF, a major peak with s20,w of 7.1 S and estimated molecular weight of 152 kDa was detected in the c(s) distribution, indicating that infliximab exists primarily in its monomeric form, in agreement with the UV-SV result. At a 10-fold molar excess of infliximab over TNF, a few peaks were detected, with the major peak at 13.9 S. In the mixtures containing infliximab:TNF at molar ratios of 2:1 and 1:1, the major peak was at 15.2 S with an apparent MW of ∼500 kDa, which suggests that this peak may correspond to (Inf)3(TNF)1 or (Inf)3(TNF)2. Similar to adalimumab, at equimolar infliximab:TNF, peaks with smaller sedimentation coefficients of 8.0 S and 12.6 S were observed. It should be noted that for each infliximab:TNF mixing ratio, the weight-average sedimentation coefficients were higher than those for adalimumab:TNF mixture ().
To assign the peaks to specific complexes, SV data for the equimolar mixture were analyzed using the hybrid local continuous distribution and global discrete species model of SEDPHAT. The 8.0 S, 12.6 S, 15.2 S, and 18.2 S peaks were designated discrete species, and the continuous distribution segment range was set at 20–100 S. Different models were generated by sequentially assigning a reasonable MW calculated for infliximab-TNF complexes with different stoichiometries to each discrete peak, using MWs of 149 kDa and 55 kDa for infliximab and TNF, respectively. The model comprising (Inf)1(TNF)1, (Inf)2(TNF)1, (Inf)3(TNF)1, and (Inf)3(TNF)2 complexes provided the best fit for the experimental databased on rmsd. Statistically indistinguishable fits with rmsd values less than the critical value were achieved using 5 other models, which are summarized in Table S3. Thus, the 8.0 S peak likely represents (Inf)1(TNF)1 and the 12.6 S peak corresponds to (Inf)2(TNF)1, which is highly consistent with the MS results (Fig. S5; ).
Finally, we performed FDS-SV on 25 nM etanercept alone and in the presence of varying concentrations of TNF (). In the absence of TNF, the sedimentation coefficient distribution had a major peak with s20,w of 5.3 S and an estimated molecular weight of 114 kDa, which was tentatively attributed to monomeric etanercept, similar to the UV-SV result. Increasing amounts of added TNF resulted in an increased 7.0 S peak area with an apparent MW of 163 kDa. Given an estimated MW of 114 kDa for the etanercept monomer and 55 kDa for the trimeric form of TNF, the 7.0 S peak probably corresponds to an (Eta)1(TNF)1 complex, consistent with previously published studies.Citation22,23 This assignment is supported by the MS results obtained for etanercept, which indicated the presence of only 1:1 complexes irrespective of TNF concentration, in contrast to adalimumab and infliximab ().
Interaction analysis in human serum
Human serum is a highly complex medium that contains a substantial amount of proteins, with human serum albumin (HSA) being the most abundant, followed by immunoglobulin (IgG). HSA is a highly soluble and stable protein that has a molecular weight of 66.5 kDa and is present in serum at a concentration of ∼40 mg/mL.Citation51 In previous studies, FDS-SV AUC experiments performed with serum alone consistently showed a peak at ∼4 S, which was attributed to HSA bound to a natively fluorescent low-molecular-weight molecule, such as flavin or heme oxidation product.Citation24 Recently, based on the fact that it has fluorescence excitation and emission spectra allowing detection with FDS, Lamola et al. concluded that this peak instead represents an oxidized bilirubin–HSA complex.Citation52 Similar results were obtained in the present study, where SV experiments performed with human serum alone revealed the presence of a peak with s20,w of ∼4.0 S. For 25 nM antagonists suspended in human serum, at an optimized gain setting, approximately half of the total fluorescent signal was due to albumin. Similar contributions from albumin were reported for 10 nM anti-IgE IgG in human serum,Citation24 whereas in another study, the HSA signal was insignificant.Citation53 The intensity of the HSA-related signal can vary due to different amounts of bilirubin–HSA complex in serum from different sources or with storage times. In general, to minimize oxidation, and hence the amount of this unwanted contaminant, it is recommended that the freshest possible serum be used in FDS-SV studies.Citation53
In the previous attempts to apply FDS-SV to proteins in serum, imperfections of the fit arising from non-ideality or other unknown origin were encountered.Citation34,38 In our study, even though the fitted curves showed some deviations early in the run, overall they matched the shape of the experimental data with considerable accuracy, and the resulting rmsd did not exceed 1% of the total loading signal (Fig. S6). The c(s) analysis of SV data acquired for 25 nM of adalimumab, infliximab, and etanercept in the presence of various amounts of TNF in human serum yielded frictional ratio values between 2.5 and 3.5, which were higher than those observed in PBS. It has been reported that larger frictional ratio values cause peak broadening.Citation45,54 Consistent with this finding, the respective derived c(s) distributions from analysis of data acquired in human serum exhibited broader peaks than in PBS.
Under conditions where no TNF was exogenously added, a major peak with s20,w of ∼7.6 S, corresponding to a monomer, is seen in the c(s) distribution obtained for adalimumab (). According to the proximity energy theory, in highly concentrated solutions containing mixtures of macromolecules, such as cell lysates or serum, proteins can experience short-range nonspecific attractive interactions leading to the formation of supramolecular assemblies.Citation55 Presumably, due to this so-called “preferential solvation” by other serum proteins, the resulting sedimentation coefficient of the adalimumab monomer in serum was slightly larger than that obtained in PBS, confirming the observations of Demeule et al.Citation24
Figure 6. FDS-SV with 25 nM adalimumab (a), infliximab (b), and etanercept (c) in the presence of varying concentrations of TNF in human serum. The c(s) distributions in the absence of TNF (black) and of the 10:1 (blue), 2:1 (green), and 1:1 (red) molar mixtures of the respective antagonist:TNF are shown.
When TNF was added to adalimumab, the resulting c(s) peaks were not baseline- resolved, which could potentially compromise the accuracy of quantification for individual adalimumab-TNF complexes (). Nevertheless, the major peaks corresponding to complexes appeared to be at s20,w values of 11.6 S and 14.8 S, similar to the results obtained in PBS. To evaluate the fractional amount of adalimumab-TNF complexes, the signal originating from the HSA-bilirubin complex as measured in SV experiments performed with human serum alone was subtracted from the total fluorescent signal obtained by integrating the area under the c(s) distribution of adalimumab-TNF mixtures. At a 10:1 adalimumab:TNF molar ratio, the adalimumab monomer fraction decreased by more than 10%. Higher concentrations of TNF resulted in a greater decrease, with less than one-third of the adalimumab present in an uncomplexed form at adalimumab:TNF molar ratio of 2:1. Similar to the results obtained in PBS, at equimolar concentrations in serum, no free monomeric adalimumab was present, and approximately a third of the HSA-corrected fluorescence signal corresponded to the 14.8 S peak, whereas higher-order complexes contributed to approximately half of the total signal intensity. In the SV experiments performed with 50 nM and 100 nM adalimumab, at a 2-fold molar excess of TNF over adalimumab, the area under the 11.6 S peak was higher, whereas the amount of higher order complexes was lower (Fig. S3C and S3D). The tendency toward forming smaller complexes when TNF was in molar excess in serum was similar to that obtained in PBS. Even though the peak positions of the major complexes were similar, weight-average s20,w values from the resulting c(s) distributions in human serum, excluding the albumin peak, were larger than those in PBS (, Table S4). The increase in weight-average s20,w values could be a consequence of “preferential solvation” of Ada-TNF complexes by serum proteins. Another possibilty is that in serum, several non-specific complexes occur due to weak binding of adalimumab to serum protein components, in addition to Ada-TNF complexes.
The monomer peak in the c(s) distribution of 25 nM infliximab in serum was markedly broader than that in PBS and slightly shifted toward larger values, with a s20,w value of 8.8 S compared with 7.1 S (). It is difficult to attribute this substantial difference solely to macromolecule-macromolecule preferential solvation, suggesting that binding of serum protein components is most likely responsible for the peak widening. SV data for infliximab-TNF mixtures in serum showed broad continuous sedimentation coefficient distributions, with no discernible complex peaks, contrary to observations in PBS. Furthermore, in contrast to adalimumab, considerably larger amounts of infliximab complexes with sedimentation coefficients above 50 S were detected (), resulting in higher weight-average s20,w values with increased TNF concentration ().
The FDS-SV analysis of etanercept in human serum was complicated by the apparent co-sedimentation of the etanercept and HSA-bilirubin complex, causing an overlap of the corresponding peaks in c(s) distributions (). To estimate the abundance of the etanercept-derived species, the HSA signal measured in serum only was subtracted from the main peak signal. In the absence of TNF, a main peak with s20,w of 4.4 S corresponding to HSA and etanercept, and a small peak at 7.4 S were observed. Similar to infliximab, the emergence of the 7.4 S-peak, which was absent in the PBS, implies possible interactions between etanercept and serum protein components. In mixtures containing TNF, the main peak corresponding to etanercept-TNF complex was located at 6.5 S and was assumed to arise from the same complex as the 7.0 S-peak detected in PBS, and therefore was attributed to the (Eta)1(TNF)1 complex. The 8.1 S-peak probably corresponds to Eta-TNF complexes of higher stoichiometry. In contrast to adalimumab and infliximab, at an equimolar ratio approximately one-fifth of the etanercept remained uncomplexed. The peak with the largest sedimnetaion coefficient was located at 10.3 S, indicating that, with the exception of the HSA-related peak, there were no substantial differences in the c(s) profiles of etanercept in PBS and human serum.
To further exclude the possibility that fluorescent labeling of antagonists may affect their interaction with TNF, we performed experiments with the same mixing concentrations using 25 nM unlabeled antagonists and 2.5–25 nM labeled TNF (Fig. S7, S8). Within the limitations imposed by the differences in the labeling efficacy, both in PBS and human serum, distributions of antagonists-TNF complexes exhibited essentially similar patterns irrespective of whether fluorescent probe was introduced into antagonist or TNF. The results confirm that labeling does not influence the interaction. In addition, the effects of incubation times on the distribution of the formed complexes were evaluated. Overnight incubation at 37°C caused the loss of fluorescent signal in mixtures formulated in PBS, presumably due to non-specific adsorption of antagonists-TNF to vial surfaces. In the case of human serum, it appears that overnight incubation induces a decrease in the weight-average sedimentation coefficient. This effect is more pronounced in adalimumab than in infliximab mixtures, whereas it is negligible in etanercept mixtures, consistent with the UV-SV results.
Adalimumab in PBS solutions containing different concentrations of human serum albumin
The distribution of adalimumab and etanercept complexes with TNF was similar in PBS and in human serum. In contrast, a significantly broader distribution, with a considerable amount of larger complexes, was observed for infliximab in human serum compared with PBS ( and ). To clarify the mechanism driving differences in complex formation among different TNF antagonists in PBS and human serum, SV experiments were conducted in serial dilutions of HSA in PBS. This approach allowed us to assess the effects of an increasingly crowded environment on antagonist-TNF complex formation in conditions similar to those in human serum, where albumin is the most abundant protein. Because co-sedimentation of HSA with etanercept complicated data interpretation, the experiments were performed using adalimumab.
As described above, a single peak with a sedimentation coefficient of 6.8 S, corresponding to monomeric adalimumab, was present in the c(s) distribution of 25 nM adalimumab in PBS without HSA (). Upon addition of 1 mg/mL HSA, a peak at 8.4 S was observed (), presumably resulting from direct interaction between the antibody and serum albumin, consistent with findings recently reported by Hill and Laue.Citation53 Increasing amounts of HSA resulted in the overall c(s20,w) distribution shift toward lower s-values, indicating non-ideal sedimentation behavior. Additionally, an HSA concentration-dependent decrease in the area under the monomer peak, with a concomitant increase in the area under the higher sedimentation coefficient peaks, further support the hypothesis of albumin-immunoglobulin interaction. Thus, at an HSA concentration of ∼18 mg/mL, the monomer peak was located at 6.1 S and comprised less than 70% of the total fluorescence intensity. The possible interaction between adalimumab and HSA did not occur due to non-specific interaction between HSA and the fluorescent probe covalently attached to adalimumab. The area under the peaks, corresponding to putative complexes formed between Alexa-Fluor 488-labeled adalimumab and HSA, decreased with increasing concertation of non-labeled adalimumab (Fig. S9A), indicating that non-labeled adalimumab competes with the labeled adalimumab for binding to HSA, which confirms that their HSA-binding abilities are similar.
Figure 7. FDS-SV analysis in serial dilutions of HSA in PBS in the absence (a) or presence of 5 mg/mL human immunoglobulin (b). The c(s) distributions of 25 nM adalimumab in 0 (black), 1 (purple), 2.5 (blue), 5 (magenta), 10 (green), and 18 mg/mL (red) HSA are shown. For the clarity of presentation, the derived c(s) distributions were normalized against the height of the main peak.
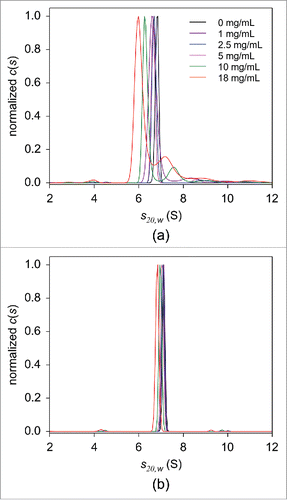
Interestingly, adalimumab in human serum (where HSA is present at a concentration of ∼40 mg/mL), other than the bilirubin-HSA peak at 4.1 S, produced a main peak at 7.6 S, which was attributed to monomeric adalimumab, and no larger peaks were detected. To explain the differences in the c(s20,w) profiles derived from HSA solutions and human serum, we performed SV experiments using HSA dilutions in the presence of human immunoglobulin at a concentration 5 mg/mL. The resulting c(s20,w) distributions revealed that the observed HSA concentration dependence was considerably diminished with the addition of immunoglobulin. Adalimumab was detected primarily in its monomeric form, indicated by the presence of the 7.0 S peak (). These results suggest that endogenous serum immunoglobulin interferes with the interaction of adalimumab and HSA.
In contrast to the c(s20,w) profiles derived from SV experiments in PBS, where the major Ada-TNF peaks at 11.9 S and 14.5 S were well-resolved (), in PBS containing 40 mg/mL HSA, broad distributions of complexes with no discernable peaks were obtained for the mixtures of 25 nM adalimumab and 0–25 nM TNF (Fig. S9B). Despite these differences, the weight-average sedimentation coefficient decreased with increasing TNF concentration irrespective of whether HSA was present (Table S5).
Measurement of FcγR activation by antagonist-TNF complexes
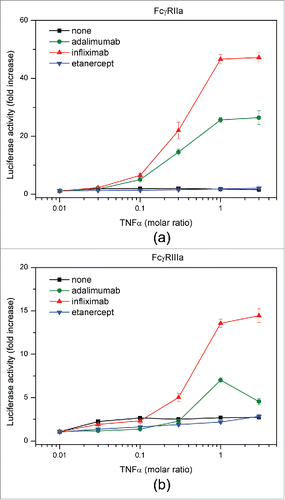
We evaluated the ability of antagonists to bind FcγRs to elucidate the effects of different complex sizes among the 3 antagonists on FcγR-induced activation of immune cells. In the absence of TNF, antagonists exhibited the same degree of FcγR binding (data not shown). We performed a cell-based reporter assay using Jurkat/FcγRIIa/NFAT-Luc and Jurkat/FcγRIIIa/NFAT-Luc cells.Citation56 Infliximab-TNF and adalimumab-TNF complexes effectively activated both cell lines in a TNF concentration-dependent manner, and the activity of infliximab was considerably higher than that of adalimumab (). In contrast, etanercept-TNF complexes caused no change in the luciferase activity of reporter cells. These results suggest that the extent of FcγR activation depends on the size of antagonist-TNF complex.
Discussion
We performed biophysical characterization of TNF-antagonist complexes for 3 antagonists, adalimumab, infliximab, and etanercept, comparing complexes formed in human serum with those formed in PBS using AUC FDS-SV. We demonstrated that complex size varies among the 3 antagonists, providing a rationale for differences in the induction of downstream signaling pathways via antagonist complex interaction with FcγRs.
In the absence of TNF, adalimumab sedimented as a monomer in both PBS and human serum with s20,w values of 6.8 S and 7.5 S, respectively. Weight-average sedimentation coefficients calculated for solutions containing adalimumab and TNF at the same concentration were higher in human serum than in PBS (). The apparent increase in protein sedimentation coefficients in highly concentrated solutions due to a preferential solvation by other macromolecules has been discussed previously.Citation55 However, our data suggest that the increased sedimentation coefficient of adalimumab in human serum instead results from interaction with the serum protein components. In PBS, the area under peaks with higher sedimentation coefficient than adalimumab monomer was larger with higher concentrations of HSA, indicating a possible interaction between adalimumab and HSA ().
Despite differences in the weight-average sedimentation coefficients, the c(s20,w) distributions in PBS and human serum exhibited similar features. A relatively broad distribution of adalimumab-TNF complexes, with the major peaks located at ∼11.9 S and 14.5 S, was seen. Based on the results of SV data analysis using the hybrid local continuous distribution and global discrete species model of SEDPHAT, these peaks were attributed to (Ada)2(TNF)1 and (Ada)3(TNF)1 complexes, respectively (). In addition, (Ada)1(TNF)1 and (Ada)3(TNF)2 complexes were identified.
Similar findings regarding the wide distribution pattern of adalimumab-TNF complexes have been previously reported. Data obtained using SEC revealed that adalimumab:TNF complexes span a wide range of molecular weights: 600–5,800 kDaCitation20 or up to 4,000 kDa.Citation21 The molecular weight of the most thermodynamically stable complex was determined to be ∼598 kDa. Therefore, it was concluded that the complex was composed of 3 adalimumab molecules and 3 TNF molecules.Citation20 However, our SV (), MS (), and simulation analysis results () do not support this stoichiometry, but rather suggest a relationship between complex size and adalimumab-TNF concentration and molar ratio. At equimolar concentration, or with TNF in excess over adalimumab, a smaller peak with s20,w of 8.2 S, which was attributed to an (Ada)1(TNF)1 complex, was revealed (, Fig. S3A). Data in show that the weight-average sedimentation coefficient is initially higher with a higher relative adalimumab concentration, and then lower at an equimolar mixing ratio. SV experiments performed with 50 nM and 100 nM adalimumab confirmed that samples containing TNF at greater than equimolar mixing ratios exhibited lower weight-average sedimentation coefficients than those below the equimolar ratio (Table S4). Furthermore, simulation data displayed a similar trend toward increased abundance of complexes of lower stoichiometry with higher TNF concentrations (). Thus, when adalimumab is in excess, the trimeric molecules of TNF are bound by more than 2 molcules of adalimumab, whereas when TNF is in excess, (Ada)1(TNF)1 complexes are detected with a corresponding decrease in the abundance of complexes with higher stoichiometries.
Native MS revealed the presence of adalimumab-TNF complexes with 1:1, 2:1, 2:2, and 3:2 stoichiometries, consistent with the AUC results (). However, the (Ada)3(TNF)1 complex to which the 14.5 S-peak in the c(s) distribution was assigned was absent. Instead, a 408,047 Da band corresponding to (Ada)2(TNF)2 was detected. As discussed above, the slight difference in complexes detected by AUC and MS is likely attributable to differences in concentration and molar ratios at which measurements were performed.
The importance of determining the binding profiles of small molecule drug candidates to serum protein components, and incorporating these analyses into drug development protocols has been emphasized.Citation57,58 Interactions with plasma proteins can have a substantial effect on the drug pharmacokinetic properties and reduce its clinical efficacy.Citation59 Conversely, the circulatory half-lives of proteins, peptide-based drugs, and small molecules can be considerably extended by association with endogenous serum albumin.Citation60,61 The neonatal Fc receptor (FcRn) plays an important role in this process by binding to drug-carrying albumin in the same way it binds to albumin and IgG, preventing their degradation via cellular recycling pathways.Citation62 The application of FDS-SV appears to be particularly useful to elucidate possible interactions of target biopharmaceuticals with serum protein components at physiologically relevant concentrations, which cannot be accounted for using other analytical techniques.
No separate peaks could be discerned in the sedimentation coefficient distribution pattern of adalimumab-TNF complexes in PBS solutions containing 40 mg/mL HSA, and it was considerably broader than that in the absence of HSA (Fig. S9). Interestingly, even though the c(s) distribution in human serum was wider than that in PBS, it was narrower than in PBS containing HSA. These observations imply that HSA may bind weakly to adalimumab-TNF complexes when present at over 1000-fold molar excess, whereas in human serum this low affinity interaction is considerably reduced. In SV experiments with adalimumab in HSA solutions, the formation of putative adalimumab-HSA complexes was abolished in the presence of human immunoglobulin. We therefore suggest that endogenous immunoglobulin in human serum is primarily responsible for the diminished adalimumab-HSA interaction.
The formation of complexes between fluorescently labeled protein OPG at picomolar concentrations and serum albumin as low as 1 mg/mL has been recently observed.Citation53 Analogous to our results, the peak corresponding to the HSA-OPG complex in HSA solutions was not detected in serum, and it was concluded that endogenous compounds in the serum compete with the OPG molecules for binding to HSA. Our data suggest that this component might be immunoglobulin, which is abundantly present in human serum. However, to our knowledge, no data showing an interaction between immunoglobulin and human serum albumin are available, and further studies are required to assess the mutual influence of endogenous proteins on binding to serum albumin.
The c(s) distribution of infliximab-TNF complexes in PBS was similar to adalimumab with regard to complex stoichiometry, but with larger weight-average sedimentation coefficients (). Our data clearly demonstrated that the complexes that are formed and their fractional abundances depended on mixing ratios (). In contrast to results previously reported by Scallon et al.,Citation23 where a single (Inf)3(TNF)1 complex was detected when TNF was in molar excess over infliximab, the additional formation of complexes with lower stoichiometries was observed in our study. The c(s) profile of infliximab-TNF complexes in human serum was markedly different than in PBS, showing a continuous distribution of complexes with no distinct peaks (). The weight-average sedimentation coefficient of infliximab complexes at each TNF molar ratio was higher than that of adalimumab ().
In contrast to adalimumab and infliximab, etanercept formed exclusively 1:1 complexes with TNF in both PBS and human serum, with only a small amount of complexes with higher binding stoichiometry detected. This finding is consistent with the previous studies where etanercept formed 1:1 complexes.Citation21-23 Overall, our data showed clear differences in the sizes of complexes formed between these 3 antagonists and TNF. Both in PBS and in human serum, the largest complexes were formed by infliximab, followed by adalimumab, whereas the smallest complexes were formed by TNF and etanercept.
Different clinical efficacy and safety profiles of the 3 different TNF antagonists have been reported, though the mechanisms responsible for these differences remain to be elucidated. A number of clinical investigations have studied the use of these TNF inhibitors, and they suggest that adalimumab has an advantage in therapeutic treatment.Citation63 Nevertheless, the incidence of ADA in psoriasis patients being treated with infliximab and adalimumab was similar.Citation13
The differences between TNF antagonists' therapeutic actions have been partly addressed in crystallographic studies. The 3-dimensional structures of infliximab-Fab-TNFCitation64 and adalimumab-Fab-TNFCitation63 complexes have been determined, and, even though it was shown that the adalimumab epitope occupies a larger area compared with infliximab, both epitopes overlap with the TNFR binding site in a similar manner. The complexes consisted of one TNF trimer bound by the 3 Fab molecules, in agreement with our AUC results (). However, the reported structural data are insufficient to explain the ability of infliximab to form larger complexes with TNF compared with adalimumab, as revealed in our study.
Because etanercept is a soluble TNFR2-Fc recombinant protein, its epitope on TNF is also located within the TNF-receptor binding site. Although 3 molcules of TNFR2 can potentially bind to a single trimeric TNF,Citation65 in etanercept, due to the structural arrangement and spatial hindrance, where the TNFR2 portion of etanercept contacts with 2 adjacent TNF protomers and the Fc portion fused to TNFR2, the remaining binding site on TNF is considered to be inaccessible to the TNFR2 portion of other etanercept molecules.Citation63 Therefore, it was concluded that one etanercept molecule binds to one trimeric TNF molecule, which is consistent with our AUC data.
In the pathogenesis of autoimmune disorders, immune complexes have been implicated in the triggering of inflammatory responses via activation of the complement pathway or through interaction with activating FcγRs expressed on immune cells.Citation66 It was suggested that larger complexes may be more effective in activating complement, leading to faster clearance of TNF from circulation. Likewise, it has been suggested that larger complexes can be cleared faster than smaller complexes via FcγRs-mediated activation of immune cells.Citation67
In an attempt to gain a better understanding of the possible correlation between anti-TNF drug safety and efficacy and sizes of the respective complexes with TNF, activation of FcγRs by adalimumab, infliximab, and etanercept in the presence of TNF was evaluated by an FcγR reporter cell assay. The results demonstrated the highest FcγR-mediated activation of the downstream immune signaling pathway occurred with infliximab, followed by adalimumab, whereas no significant increase in activation of FcγR could be observed for etanercept (). We suggest that enhancement of the FcγR-mediated signal transduction can be attributed to a size-dependent increase in binding of mAb-TNF complexes to FcγRs, which results in more efficient clustering of the receptors. It has been shown that in the absence of exogenous TNF, adalimumab, infliximab, and etanercept were only capable of low-level binding to the activating receptors FcγRIIa and FcγRIIIa. Upon addition of TNF, however, increased binding of mAbs, but not etanercept was observed, which was suggested to be mainly due to the formation of large protein complexes by the mAbs. Consistent with these in vitro observations, adalimumab and infliximab, but not etanercept, induced antibody-dependent cell-meditated cytotoxicity.Citation68
Increased activation of FcγR leads to faster clearance of both antigens and antibodies by enhanced uptake and breakdown of the complexes. Immune cells that express activating FcγR, such as monocytes and macrophages, produce proinflammatory cytokines, particularly interferon-γ, which induces the expression of class II MHC that can, in turn, increase the presentation of the fragments of drug antibody and stimulate ADA generation. Thus, the shorter serum half-life of infliximab, as well as the higher incidence of ADA formation compared with adalimumab, can be explained by the formation of larger complexes, although further studies may be needed to increase the generalizability of these findings.
Self-aggregation is one of the major concerns with regard to the clinical safety of biological therapeutics.Citation69,70 Because the aggregated forms of mAbs resemble immune complexes, they may play an important role in inducing ADA formation.Citation71 Correlation between sizes of aggregates and immunogenicity have been considered, and larger micron-sized aggregates, often referred to as sub-visible particles, were suggested to have a higher propensity to induce immune response than nanosized aggregates.Citation72 Using the Stokes-Einstein relationship, we estimated the hydrodynamic radii of the peaks present in the c(s) distribution of adalimumab and infliximab in human serum and concluded that the soluble antagonist-TNF complexes were smaller than 100 nm. Nevertheless, our cell-reporter assay clearly showed that these complexes were able to induce considerable FcγR activation, suggesting a potential role in immunogenicity and manifestation of adverse effects.
In conclusion, the results reported here provide clear evidence that FDS-SV is an extremely valuable technique for the prediction of therapeutic action and in-depth understanding of the binding of target protein drugs to their respective antigens directly in human serum at physiologically relevant concentrations. It is anticipated that incorporating FDS-AUC in the drug development process will positively contribute to optimal design of pharmaceuticals with enhanced efficacy and minimized immunogenicity.
Materials and methods
Proteins
Adalimumab (Humira®) was purchased from Eisai, infliximab (Remicade®) was obtained from Mitsubishi Tanabe Pharma, and etanercept (Enbrel®) was received from Takeda Pharmaceutical Company Limited. Recombinant human soluble TNF was produced recombinantly using a silkworm baculovirus expression system by ProCube® Biotechnology Center at the Sysmex Research and Development Center in Kobe. Recombinant human TNF purified from E coli was purchased from R&D Systems Inc. FDS-SV experiments performed with adalimumab and TNF prepared using a baculovirus and an E coli expression system yielded essentially the same results (Fig. S3 and S10) and therefore, unless otherwise stated, baculovirus-expressed TNF was used in all experiments.
Methods
Absorbance-detected sedimentation velocity analytical ultracentrifugation
Two μM respective antagonist and 2 μM TNF were mixed in PBS pH 7.4 (Gibco) and incubated at 20°C for 2 hours or at 37°C overnight before AUC. Sedimentation velocity (SV) experiments with the resulting mixtures and with 2 μM antagonists in the absence of TNF were performed using an Optima XL-I analytical ultracentrifuge equipped with absorbance optics (Beckman Coulter) at rotor speed of 42,000 rpm. Scans were acquired at a wavelength of 280 nm with 30-μm radial increments. Experiments with 2 μM TNF were conducted at a rotor speed of 60,000 rpm using absorbance detection at 230 nm.
Dynamic light scattering
DLS was conducted at 25°C using ZetaSizer Nano (Malvern). Adalimumab, infliximab, and etanercept were diluted to 3.3 μM using PBS, filtered by 0.22 μm-pore size syringe filter (Millipore), and mixtures containing the respective antagonist:TNF at molar ratios of 1:0, 1:0.3, and 1:1 were prepared.
Protein labeling
Labeling of target proteins with a fluorescent probe that can be excited using 488-nm laser light and which emission wavelengths are greater than 505 nm is necessary for FDS measurements. We chose Alexa Fluor® 488 fluorescent dye (Molecular Probes®), which meets these requirements, and is also relatively insensitive to pH, and highly resistant to photobleaching. In addition, it was shown to be stable during AUC experiments conducted in serum.Citation24,53 Labeling of adalimumab, infliximab, etanercept, and TNF was performed according to the following protocol. Each TNF antagonist was at first diluted to 15 mg/mL to a final volume of 100 μL. Then, 10 μL of 1 M sodium bicarbonate (pH 8.3) and 2.2 μL of 10 mg/mL Alexa Fluor® 488 stock solution (5 mg reactive dye/0.5 mL dimethyl formamide) were added and the resulting mixture was allowed to react for 4 h at room temperature. The reaction was quenched with 10 μL of 1.5 M hydroxylamine (pH 8.5) and mixture was allowed to react for another 1 h at room temperature. The purification of Alexa-labeled antagonists was performed by SEC using Alliance 1100 HPLC system with TSKG3000 SWXL column and 1× PBS pH 7.4 as the mobile phase. The final solution concentrations were 1.8, 1.4, 2.5, and 1.2 μM for adalimumab, infliximab, etanercept, and TNF, respectively.
Fluorescence-detected sedimentation velocity analytical ultracentrifugation
Twenty-five nM respective labeled antagonist and 0–25 nM TNF were mixed in PBS or human serum (MP Biomedicals) and incubated at 20°C for 2 hours before FDS-SV AUC. Similar mixtures containing 25 nM unlabeled antagonist and 2.5–25 nM labeled TNF were incubated at 20°C for 2 hours or at 37°C overnight before FDS-SV AUC. Additionally, mixtures containing 50 nM and 100 nM labeled adalimumab were analyzed. The experiments were performed using fluorescence optics (Aviv Biomedical) at a rotor speed of 42,000 rpm. To prevent non-specific adsorption of protein to centerpiece walls and windows, 0.1 mg/mL lysozyme was added to PBS.Citation34 To clarify the mechanism driving differences in complex formation among different TNF antagonists in PBS and human serum, SV experiments were conducted in serial dilutions of pooled human serum albumin in PBS.
The acquired data were analyzed with SEDFIT (version 14.6e) using the continuous c(s) distribution model.Citation39,73 The apparent sedimentation coefficients were converted to s20,w using density and viscosity of the respective buffer solutions measured using an Anton Paar density meter DMA4500 and viscometer Lovis 2000ME, respectively. To evaluate the fractional amount of antagonist-TNF complexes, the signal originating from the HSA-bilirubin complex as measured in SV experiments performed with human serum alone was subtracted from the total fluorescent signal obtained by integrating the area under the c(s) distribution of antagonist-TNF mixtures.
Mass spectrometry under non-denaturing conditions
TNF, adalimumab, and infliximab were buffer-exchanged into 150 mM ammonium acetate, pH 6.8 by passing the proteins through a Bio-Spin 6 column (Bio-Rad). The buffer-exchanged TNF (2 μM trimer) and respective antagonist (2, 2.5, 3, and 4 μM) were mixed, incubated at 25°C for 20 min, and analyzed by nanoflow electrospray ionization MS using gold-coated glass capillaries made in-house (∼2–5 µL sample loaded per analysis). Spectra were recorded on a SYNAPT G2-Si HDMS mass spectrometer (Waters Corporation) in positive ionization mode at 1.33 kV with a 150 V sampling cone voltage and source offset voltage, 0 V trap and transfer collision energy, and 5 mL/min trap gas flow. The spectra were calibrated using 1 mg/mL cesium iodide and analyzed using MassLynx software (Waters Corporation).
Isothermal titration calorimetry
ITC experiments were performed with a Fab fragment and full-length adalimumab at 25°C using an iTC200 (GE Healthcare) or PEAQ-ITC (Malvern) instruments. Prior to each experiment, TNF, adalimumab-Fab, and full-length adalimumab were extensively dialyzed against PBS containing 0.1% polysorbate 80. For adalimumab-Fab binding to TNF, 9.8 μM adalimumab-Fab was placed into the cell and 39.8 μM trimeric TNF was loaded into the syringe. For the experiments with full-length adalimumab, 1.7 μM adalimumab was placed into the cell and the 27.5 μM TNF was loaded into the syringe. Titrations consisted of a preliminary 1-μL injection, followed by 2-μL injections with a 120 s interval between 2 successive injections. The integration of thermograms was performed using NITPIC softwareCitation74 and the resulting isotherms were fit to a A+B+B+B AB+B+B ⇌ ABB+B ⇌ ABBB binding model using the SEDPHAT software.
FcγR reporter assay
Stable cell lines Jurkat/FcγRIIa/NFAT-Luc and Jurkat/FcγRIIIa/NFAT-Luc expressing FcγRIIa or FcγRIIIa, respectively, were established as described in.ref. Citation56 TNF antagonists were mixed with TNF at different molar ratio and incubated for 30 mins at 37°C. Mixtures of respective antagonist and TNF were added to Jurkat/FcγRs/NFAT-Luc cells to the final concentration of antagonist of 1 μg/mL. After incubation for 4 hr at 37°C in 5% CO2, luciferase activities were measured by using ONE-Glo Luciferase Assay System (Promega).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
M.K. expressed and purified recombinant TNF. M.N. performed labeling. E.K. performed AUC. M.T., T.S., and A.I.-W. performed DLS. K.I. performed native MS. T.M. performed ITC. M.T., T.S., A.I.-W., and H.W. performed reporter assays. S.U. directed the study. E.K. wrote the manuscript with assistance from all other authors.
Supplemental_Material.docx
Download MS Word (10.2 MB)Acknowledgment
We thank Ms. Masami Yokoyama for technical assistance.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan under grants No. 15K14457, 16H00770, 26102530 (S.U.), 24790183 (M.T.), and 20590167 (A.I.-W); Okazaki ORION project (S.U.); Joint Studies Program in the Okazaki BIO-NEXT project of the Okazaki Institute for Integrative Bioscience (S.U.); “Research on Regulatory Harmonization and Evaluation of Pharmaceuticals, Medical Devices, Regenerative and Cellular Therapy Products, Gene Therapy Products, and Cosmetics” from Japan Agency for Medical Research and development (AMED) (M.T.)
References
- Salfeld J, Kaymakçalan Z, Tracey D, Roberts A, Kamen R. Generation of fully human anti-TNF antibody D2E7. Arthritis Rheum 1998; 41:S57.
- AbbVie Corporation, St-Laurent QC. Humira (adalimumab): Product monograph; 2015 Nov 15 [accessed 2016 Oct 19]. http://www.abbvie.ca/content/dam/abbviecorp/ca/english/docs/HUMIRA_PM_EN.pdf.
- Knight DM, Trinh H, Le J, Siegel S, Shealy D, McDonough M, Scallon B, Moore MA, Vilcek J, Daddona P, et al. Construction and initial characterization of a mouse-human chimeric anti-TNF antibody. Mol Immunol 1993; 30(16):1443-53; PMID:8232330; http://dx.doi.org/10.1016/0161-5890(93)90106-L
- Janssen Inc., Toronto, Ontario. Remicade (infliximab): Product monograph; 2015 Jul 22 [accessed 2016 Oct 19]. https://www.janssen.com/canada/sites/www_janssen_com_canada/files/product/pdf/rmc07222015cpm_nc_183500.pdf.
- Immunex Corporation, Thousand Oaks, CA. Enbrel (etanercept): Product monograph; 2015 Oct 19 [accessed 2016 Oct 19]. https://www.amgen.ca/Enbrel_PM.pdf.
- Moreland LW, Baumgartner SW, Schiff MH, Tindall EA, Fleischmann RM, Weaver AL, Ettlinger RE, Cohen S, Koopman WJ, Mohler K, et al. Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)-Fc fusion protein. N Engl J Med 1997; 337(3):141-7; PMID:9219699; http://dx.doi.org/10.1056/NEJM199707173370301
- Langley RG, Gupta AK, Cherman AM, Inniss KA. Biologic therapeutics in the treatment of psoriasis. Part 1: review. J Cutan Med Surg 2007; 11(3):99-122.
- van der Heijde D, Kivitz A, Schiff MH, Sieper J, Dijkmans BA, Braun J, Dougados M, Reveille JD, Wong RL, Kupper H, et al. Efficacy and safety of adalimumab in patients with ankylosing spondylitis: results of a multicenter, randomized, double-blind, placebo-controlled trial. Arthritis Rheum 2006; 54(7):2136-46; PMID:16802350; http://dx.doi.org/10.1002/art.21913
- Hanauer SB, Sandborn WJ, Rutgeerts P, Fedorak RN, Lukas M, MacIntosh D, Panaccione R, Wolf D, Pollack P. Human anti-tumor necrosis factor monoclonal antibody (adalimumab) in Crohn's disease: the CLASSIC-I trial. Gastroenterology 2006; 130(2):323-33; quiz 591; PMID:16472588; http://dx.doi.org/10.1053/j.gastro.2005.11.030
- Kestens C, van Oijen MG, Mulder CL, van Bodegraven AA, Dijkstra G, de Jong D, Ponsioen C, van Tuyl BA, Siersema PD, Fidder HH, et al. Adalimumab and infliximab are equally effective for Crohn's disease in patients not previously treated with anti-tumor necrosis factor-alpha agents. Clin Gastroenterol Hepatol 2013; 11(7):826-31; PMID:23376000; http://dx.doi.org/10.1016/j.cgh.2013.01.012
- Osterman MT, Haynes K, Delzell E, Zhang J, Bewtra M, Brensinger C, Chen L, Xie F, Curtis JR, Lewis JD. Comparative effectiveness of infliximab and adalimumab for Crohn's disease. Clin Gastroenterol Hepatol 2014; 12(5):811-7 e3; PMID:23811254; http://dx.doi.org/10.1016/j.cgh.2013.06.010
- Scheinfeld N. A comprehensive review and evaluation of the side effects of the tumor necrosis factor alpha blockers etanercept, infliximab and adalimumab. J Dermatolog Treat 2004; 15(5):280-94; PMID:15370396; http://dx.doi.org/10.1080/09546630410017275
- Hsu L, Armstrong AW. Anti-drug antibodies in psoriasis: a critical evaluation of clinical significance and impact on treatment response. Expert Rev Clin Immunol 2013; 9(10):949-58; PMID:24128157; http://dx.doi.org/10.1586/1744666X.2013.836060
- Shukla R, Vender RB. Pharmacology of TNF inhibitors. TNF-alpha Inhibitors 2006:23-44; http://dx.doi.org/10.1007/3-7643-7438-1_3
- Aarden L, Ruuls SR, Wolbink G. Immunogenicity of anti-tumor necrosis factor antibodies-toward improved methods of anti-antibody measurement. Curr Opin Immunol 2008; 20(4):431-5; PMID:18619538; http://dx.doi.org/10.1016/j.coi.2008.06.011
- Lombardi G, Perego S, Sansoni V, Diani M, Banfi G, Altomare G. Anti-adalimumab antibodies in psoriasis: lack of clinical utility and laboratory evidence. BMJ Open 2016; 6(12):e011941; PMID:27940624; http://dx.doi.org/10.1136/bmjopen-2016-011941
- Kaymakcalan Z, Sakorafas P, Bose S, Scesney S, Xiong L, Hanzatian DK, Salfeld J, Sasso EH. Comparisons of affinities, avidities, and complement activation of adalimumab, infliximab, and etanercept in binding to soluble and membrane tumor necrosis factor. Clin Immunol 2009; 131(2):308-16; PMID:19188093; http://dx.doi.org/10.1016/j.clim.2009.01.002
- Schroter C, Gunther R, Rhiel L, Becker S, Toleikis L, Doerner A, Becker J, Schönemann A, Nasu D, Neuteboom B, et al. A generic approach to engineer antibody pH-switches using combinatorial histidine scanning libraries and yeast display. MAbs 2015; 7(1):138-51; PMID:25523975; http://dx.doi.org/10.4161/19420862.2014.985993
- Ogura T, Tanaka Y, Toyoda H. Whole cell-based surface plasmon resonance measurement to assess binding of anti-TNF agents to transmembrane target. Anal Biochem 2016; 508:73-7; PMID:27349512; http://dx.doi.org/10.1016/j.ab.2016.06.021
- Santora LC, Kaymakcalan Z, Sakorafas P, Krull IS, Grant K. Characterization of noncovalent complexes of recombinant human monoclonal antibody and antigen using cation exchange, size exclusion chromatography, and BIAcore. Anal Biochem 2001; 299(2):119-29; PMID:11730333; http://dx.doi.org/10.1006/abio.2001.5380
- Kohno T, Tam LT, Stevens SR, Louie JS. Binding characteristics of tumor necrosis factor receptor-Fc fusion proteins vs anti-tumor necrosis factor mAbs. J Investig Dermatol Symp Proc 2007; 12(1):5-8; PMID:17502862;http://dx.doi.org/10.1038/sj.jidsymp.5650034
- Kim MS, Lee SH, Song MY, Yoo TH, Lee BK, Kim YS. Comparative analyses of complex formation and binding sites between human tumor necrosis factor-alpha and its three antagonists elucidate their different neutralizing mechanisms. J Mol Biol 2007; 374(5):1374-88; PMID:17996896; http://dx.doi.org/10.1016/j.jmb.2007.10.034
- Scallon B, Cai A, Solowski N, Rosenberg A, Song XY, Shealy D, Wagner C. Binding and functional comparisons of two types of tumor necrosis factor antagonists. J Pharmacol Exp Ther 2002; 301(2):418-26; PMID:11961039; http://dx.doi.org/10.1124/jpet.301.2.418
- Demeule B, Shire SJ, Liu J. A therapeutic antibody and its antigen form different complexes in serum than in phosphate-buffered saline: a study by analytical ultracentrifugation. Anal Biochem 2009; 388(2):279-87; PMID:19289095; http://dx.doi.org/10.1016/j.ab.2009.03.012
- Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Protein Sci 2013; Chapter 20: 20.12.1-20.12.49; Unit20 12; PMID:23377850; http://dx.doi.org/10.1002/0471140864.ps2012s71
- Chaton CT, Herr AB. Elucidating complicated assembling systems in biology using size-and-shape analysis of sedimentation velocity data. Methods Enzymol 2015; 562:187-204; PMID:26412652; http://dx.doi.org/10.1016/bs.mie.2015.04.004
- Liu J, Yadav S, Andya J, Demeule B, Shire SJ. Analytical ultracentrifugation and its role in development and research of therapeutical proteins. Methods Enzymol 2015; 562:441-76; PMID:26412663; http://dx.doi.org/10.1016/bs.mie.2015.04.008
- Krayukhina E, Uchiyama S. Analytical ultracentrifugation. In: Senda T, Maenaka K, eds. Advanced Methods in Structural Biology: Springer Japan, 2016:165-83; http://dx.doi.org/10.1007/978-4-431-56030-2_10
- Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci 2002; 11(9):2067-79; PMID:12192063; http://dx.doi.org/10.1110/ps.0207702
- Gabrielson JP, Brader ML, Pekar AH, Mathis KB, Winter G, Carpenter JF, Randolph TW. Quantitation of aggregate levels in a recombinant humanized monoclonal antibody formulation by size-exclusion chromatography, asymmetrical flow field flow fractionation, and sedimentation velocity. J Pharm Sci 2007; 96(2):268-79; PMID:17080424; http://dx.doi.org/10.1002/jps.20760
- Philo JS. A critical review of methods for size characterization of non-particulate protein aggregates. Curr Pharm Biotechnol 2009; 10(4):359-72; PMID:19519411; http://dx.doi.org/10.2174/138920109788488815
- den Engelsman J, Garidel P, Smulders R, Koll H, Smith B, Bassarab S, Seidl A, Hainzl O, Jiskoot W. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm Res 2011; 28(4):920-33; PMID:20972611; http://dx.doi.org/10.1007/s11095-010-0297-1
- MacGregor IK, Anderson AL, Laue TM. Fluorescence detection for the XLI analytical ultracentrifuge. Biophys Chem 2004; 108(1-3):165-85; PMID:15043928; http://dx.doi.org/10.1016/j.bpc.2003.10.018
- Kingsbury JS, Laue TM. Fluorescence-detected sedimentation in dilute and highly concentrated solutions. Methods Enzymol 2011; 492:283-304; PMID:21333796; http://dx.doi.org/10.1016/B978-0-12-381268-1.00021-5
- Zhao H, Mayer ML, Schuck P. Analysis of protein interactions with picomolar binding affinity by fluorescence-detected sedimentation velocity. Anal Chem 2014; 86(6):3181-7; PMID:24552356; http://dx.doi.org/10.1021/ac500093m
- Nelson TG, Ramsay GD, Perugini MA. Fluorescence Detection System. In: Uchiyama S, Arisaka F, Stafford WF, Laue T, eds. Analytical Ultracentrifugation: Springer Japan, 2016:39-61; http://dx.doi.org/10.1007/978-4-431-55985-6_4
- Cole JW, Lary JW, Moody TP, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. In: Correia JJ, Detrich H, eds. Methods Cell Biol. San Diego: Elsevier, 2008:143-79; PMID:17964931; http://dx.doi.org/10.1016/S0091-679X(07)84006-4
- Kroe RR, Laue TM. NUTS and BOLTS: applications of fluorescence-detected sedimentation. Anal Biochem 2009; 390(1):1-13; PMID:19103145; http://dx.doi.org/10.1016/j.ab.2008.11.033
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 2000; 78(3):1606-19; PMID:10692345; http://dx.doi.org/10.1016/S0006-3495(00)76713-0
- Schuck P. Diffusion-deconvoluted sedimentation coefficient distributions for the analysis of interacting and non-interacting protein mixtures. In: Scott DJ, Harding SE, Rowe AJ, eds. Analytical Ultracentrifugation: Techniques and Methods. UK: RSC Publishing, 2005:26-49; http://dx.doi.org/10.1039/9781847552617-00026
- Eck MJ, Sprang SR. The structure of tumor necrosis factor-alpha at 2.6-a resolution - implications for receptor-binding. J Biol Chem 1989; 264(29):17595-605; PMID:2551905
- Narhi LO, Philo JS, Li T, Zhang M, Samal B, Arakawa T. Induction of alpha-helix in the beta-sheet protein tumor necrosis factor-alpha: acid-induced denaturation. Biochemistry 1996; 35(35):11454-60; PMID:8784201; http://dx.doi.org/10.1021/bi952767n
- Houel S, Hilliard M, Yu YQ, McLoughlin N, Martin SM, Rudd PM, Williams JP, Chen W. N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Analytical Chem 2014; 86(1):576-84; PMID:24308717; http://dx.doi.org/10.1021/ac402726h
- Lyons DF, Lary JW, Husain B, Correia JJ, Cole JL. Are fluorescence-detected sedimentation velocity data reliable? Anal Biochem 2013; 437(2):133-7; PMID:23499970; http://dx.doi.org/10.1016/j.ab.2013.02.019
- Krayukhina E, Uchiyama S, Fukui K. Effects of rotational speed on the hydrodynamic properties of pharmaceutical antibodies measured by analytical ultracentrifugation sedimentation velocity. Eur J Pharm Sci 2012; 47(2):367-74; PMID:22728396; http://dx.doi.org/10.1016/j.ejps.2012.06.005
- Oda M, Uchiyama S, Robinson CV, Fukui K, Kobayashi Y, Azuma T. Regional and segmental flexibility of antibodies in interaction with antigens of different size. FEBS J 2006; 273(7):1476-87; PMID:16689933; http://dx.doi.org/10.1111/j.1742-4658.2006.05168.x
- Oda M, Uchiyama S, Noda M, Nishi Y, Koga M, Mayanagi K, Azuma T. Effects of antibody affinity and antigen valence on molecular forms of immune complexes. Mol Immunol 2009; 47(2-3):357-64; PMID:19800690; http://dx.doi.org/10.1016/j.molimm.2009.09.009
- Noda M, Uchiyama S, McKay AR, Morimoto A, Misawa S, Yoshida A, Shimahara H, Takinowaki H, Nakamura S, Kobayashi Y, et al. Assembly states of the nucleosome assembly protein 1 (NAP-1) revealed by sedimentation velocity and non-denaturing MS. Biochem J 2011; 436(1):101-12; PMID:21348863; http://dx.doi.org/10.1042/BJ20102063
- Ishii K, Noda M, Yagi H, Thammaporn R, Seetaha S, Satoh T, Kato K, Uchiyama S. Disassembly of the self-assembled, double-ring structure of proteasome alpha7 homo-tetradecamer by alpha6. Sci Rep 2015; 5:18167; PMID:26657688; http://dx.doi.org/10.1038/srep18167
- Kemmer G, Keller S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat Protoc 2010; 5(2):267-81; PMID:20134427; http://dx.doi.org/10.1038/nprot.2009.182
- Peters T. Metabolism: Albumin in the body. In: All about albumin: Biochemistry, genetics and medical applications. Academic Press, 1995; 188-250.
- Lamola AA, Russo M. Fluorescence excitation spectrum of bilirubin in blood: a model for the action spectrum for phototherapy of neonatal jaundice. Photochem Photobiol 2014; 90(2):294-6; PMID:23998276; http://dx.doi.org/10.1111/php.12167
- Hill JJ, Laue TM. Protein Assembly in Serum and the Differences from Assembly in Buffer. Methods Enzymol 2015; 562:501-27; PMID:26412665; http://dx.doi.org/10.1016/bs.mie.2015.06.012
- Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J 2002; 82(2):1096-111; PMID:11806949; http://dx.doi.org/10.1016/S0006-3495(02)75469-6
- Laue T. Proximity energies: a framework for understanding concentrated solutions. J Mol Recognit 2012; 25(3):165-73; PMID:22407980; http://dx.doi.org/10.1002/jmr.2179
- Tada M, Ishii-Watabe A, Suzuki T, Kawasaki N. Development of a cell-based assay measuring the activation of FcgammaRIIa for the characterization of therapeutic monoclonal antibodies. PLoS One 2014; 9(4):e95787; PMID:24752341; http://dx.doi.org/10.1371/journal.pone.0095787
- Banker MJ, Clark TH. Plasma/serum protein binding determinations. Curr Drug Metab 2008; 9(9):854-9; PMID:18991581; http://dx.doi.org/10.2174/138920008786485065
- Howard ML, Hill JJ, Galluppi GR, McLean MA. Plasma protein binding in drug discovery and development. Comb Chem High Throughput Screen 2010; 13(2):170-87; PMID:20053162; http://dx.doi.org/10.2174/138620710790596745
- Baker MA, Schneider EK, J XH, Cooper MA, Li J, Velkov T. The plasma protein binding proteome of ertapenem: A novel compound-centric proteomic approach for elucidating drug-plasma protein binding interactions. ACS Chem Biol 2016; 11(12):3353-64; PMID:27682196; http://dx.doi.org/10.1021/acschembio.6b00700
- Sleep D. Albumin and its application in drug delivery. Expert Opin Drug Deliv 2015; 12(5):793-812; PMID:25518870; http://dx.doi.org/10.1517/17425247.2015.993313
- Sand KM, Bern M, Nilsen J, Noordzij HT, Sandlie I, Andersen JT. Unraveling the interaction between fcrn and albumin: opportunities for design of albumin-based therapeutics. Front Immunol 2014; 5:682; PMID:25674083; http://dx.doi.org/10.3389/fimmu.2014.00682
- Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, Anderson CL. The major histocompatibility complex–related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med 2003; 197(3):315-22; PMID:12566415; http://dx.doi.org/10.1084/jem.20021829
- Hu S, Liang S, Guo H, Zhang D, Li H, Wang X, Yang W, Qian W, Hou S, Wang H, et al. Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view. J Biol Chem 2013; 288(38):27059-67; PMID:23943614; http://dx.doi.org/10.1074/jbc.M113.491530
- Liang S, Dai J, Hou S, Su L, Zhang D, Guo H, Lou Z. Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab. J Biol Chem 2013; 288(19):13799-807; PMID:23504311; http://dx.doi.org/10.1074/jbc.M112.433961
- Mukai Y, Nakamura T, Yoshikawa M, Yoshioka Y, Tsunoda S, Nakagawa S, Yamagata Y, Tsutsumi Y. Solution of the structure of the TNF-TNFR2 complex. Sci Signal 2010; 3(148):ra83; PMID:21081755; http://dx.doi.org/10.1126/scisignal.2000954
- Ravetch JV, Clynes RA. Divergent roles for Fc receptors and complement in vivo. Annu Rev Immunol 1998; 16(16):421-32; PMID:9597136; http://dx.doi.org/10.1146/annurev.immunol.16.1.421
- Krishna M, Nadler SG. Immunogenicity to biotherapeutics - the role of anti-drug immune complexes. Front Immunol 2016; 7:21; PMID:26870037; http://dx.doi.org/10.3389/fimmu.2016.00021
- Arora T, Padaki R, Liu L, Hamburger AE, Ellison AR, Stevens SR, Louie JS, Kohno T. Differences in binding and effector functions between classes of TNF antagonists. Cytokine 2009; 45(2):124-31; PMID:19128982; http://dx.doi.org/10.1016/j.cyto.2008.11.008
- Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J 2006; 8(3):E501-7; PMID:17025268; http://dx.doi.org/10.1208/aapsj080359
- Krayukhina E, Uchiyama S, Nojima K, Okada Y, Hamaguchi I, Fukui K. Aggregation analysis of pharmaceutical human immunoglobulin preparations using size-exclusion chromatography and analytical ultracentrifugation sedimentation velocity. J Biosci Bioeng 2013; 115(1):104-10; PMID:22925901; http://dx.doi.org/10.1016/j.jbiosc.2012.07.021
- Jefferis R. Posttranslational modifications and the immunogenicity of biotherapeutics. J Immunol Res 2016; 2016:5358272; PMID:27191002; http://dx.doi.org/10.1155/2016/5358272
- Moussa EM, Panchal JP, Moorthy BS, Blum JS, Joubert MK, Narhi LO, Topp EM. Immunogenicity of therapeutic protein aggregates. J Pharm Sci 2016; 105(2):417-30; PMID:26869409; http://dx.doi.org/10.1016/j.xphs.2015.11.002
- Brautigam CA. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol 2015; 562:109-33; PMID:26412649; http://dx.doi.org/10.1016/bs.mie.2015.05.001
- Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem 2012; 84(11):5066-73; PMID:22530732; http://dx.doi.org/10.1021/ac3007522