ABSTRACT
Tandem single-chain variable fragment (scFv) bispecific antibodies (bsAb) are one of the most promising bsAb formats reported thus far. Yet, because of their increased aggregation propensity, high impurity content due to low expression level, smaller size and lack of the Fc region, it is challenging to isolate these products with high yield and purity within a limited number of purification steps in a scalable fashion. A robust purification process that is able to circumvent these issues is therefore of critical importance to allow effective isolation of this group of antibodies. We investigated the addition of sodium chloride (NaCl), calcium chloride (CaCl2), and L-arginine monohydrochloride (Arg·HCl) to the elution buffer of Protein L affinity chromatography, and propose here a novel mechanism for the modification of Protein L binding avidity that can lead to enhanced high molecular weight (HMW)-monomer separation, a preferential strengthening effect of the HMW-Protein L interaction compared to the monomer-Protein L interaction. In particular, we found Arg·HCl to be the most effective salt additive in terms of purity and recovery. The mechanism we propose is different from the widely reported chaotropic effect exerted by salt additives observed in Protein A chromatography. We also demonstrate here that a final eluate containing <1% HMW species and <100 ppm host cell proteins can be obtained within a two-step process with an overall yield of 65%, highlighting the promising suitability of Protein L affinity chromatography for the purification of kappa light chain-containing tandem scFv bsAb.
Introduction
Bispecific antibodies (bsAbs) recognize and bind to two different epitopes, bringing them into close proximity, thus allowing them to display novel functionalities otherwise absent in their parental antibodies. This dual-targeting concept has yielded remarkably promising clinical results, as exemplified by the 2 bsAbs (blinatumomabCitation1,Citation2 and emicizumabCitation3) that are currently marketed and the substantial number (over 85) that are in clinical development.Citation4 The enormous therapeutic potential of bispecific antibodies, particularly in the treatment of cancer and inflammatory disorders, has led to the development of many different formats of recombinant bsAbs.Citation4–Citation9 Notably, a particular format of bsAbs that has attracted considerable interest is that of tandem single-chain variable fragments (scFv), which comprise the fusion of two scFv regions, often connected by a short peptide linker.Citation4–Citation9 Blinatumomab, the bispecific T-cell engager (BiTE®) currently marketed as a treatment for acute lymphoblastic leukemia, is an example of a tandem scFv bsAb, consisting of two scFv regions linked by a glycine-serine peptide, targeting the CD3 antigen present on cytotoxic T-cells and CD19 antigen on B lymphocytes ().Citation1,
Figure 1. Schematic representation of the structures of monoclonal antibodies (a) and tandem scFv bsAbs (b). A specific example of a tandem scFv bsAb is blinatumomab, with two different scFv fragments that bind to the CD3 and B lymphocyte antigen CD19, respectively, and its biosimilar was used as a model tandem scFv bsAb molecule in this study
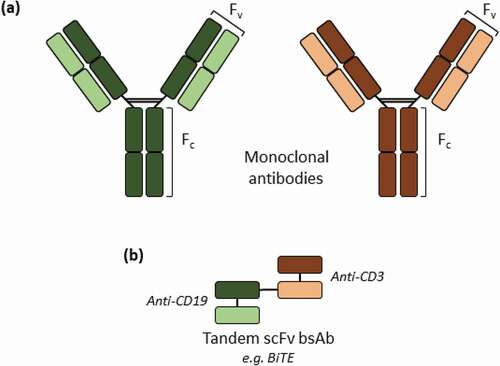
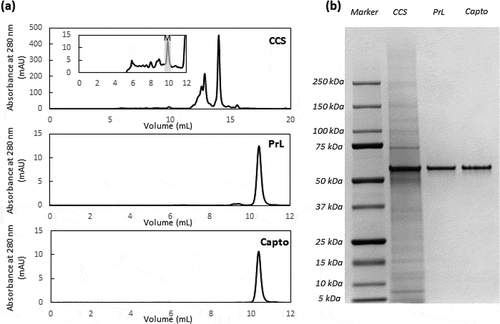
Figure 2. AKTA chromatogram of TOYOPEARLTM AF-rProtein L-650F with a step elution at pH 3.0, with the inset illustrating the HPLC-SEC chromatogram of the whole peak collected from TOYOPEARLTM AF-rProtein L-650F chromatography
In comparison to the rapid advances in cell line development for tandem scFv bsAb, relatively few publications describe the scalable purification of these antibodies. As tandem scFv bsAb lack the Fc region, well-established purification protocols commonly used for the purification of monoclonal IgG, such as Protein A affinity chromatography, cannot be used for the purification of various tandem scFv bsAb, particularly those that are not derived from the VH3 gene family.Citation10 Furthermore, the absence of the Fc region has been reported to render the antibody more aggregation-prone compared to the parental conventional immunoglobulins.Citation11,Citation12 The overall small sizes and high impurity content due to low expression levels of this particular type of bsAb also pose additional challenges in the downstream purification process, which should yield products of high purity within a limited number of purification steps. A commonly used method for the purification of tandem scFv bsAb reported in literatureCitation13-Citation18 involves the use of immobilized metal affinity chromatography (IMAC), where the target molecule is engineered to contain a poly-histidine tag that is able to bind to immobilized metal ions. Consequently, an additional proteolytic digest step to remove this poly-histidine tag is preferentially performed at the end of the purification process during therapeutic drug development. Coupled with size exclusion chromatography (SEC) as a second purification step, a purity of >95%, as estimated by SDS-PAGE gels, have frequently been reported.Citation16,Citation18 The use of SEC is, however, limited to purification processes at the laboratory scale due to scalability issues.
Another alternative purification method for the purification of tandem scFv BsAb is the use of Protein L affinity chromatography,Citation19,Citation20 which eliminates the need for the presence of a poly-histidine tag on the target molecule. Isolated from the bacteria Peptostreptococcus magnus, Protein L is a cell surface protein that interacts with the variable region of the antibody’s kappa (К) light chain, in particular, the К1, К3, and К4 light chains, allowing it to bind to approximately 67% of human immunoglobulins and 99% of mouse immunoglobulins.Citation21–Citation23 So far, the use of Protein L affinity chromatography has been reported for research purposes only, with overall purity reported as 50 – 70% after a single affinity chromatographic stepCitation20 and requiring additional Protein A or Protein G chromatographic steps and SEC to achieve higher purity.Citation19
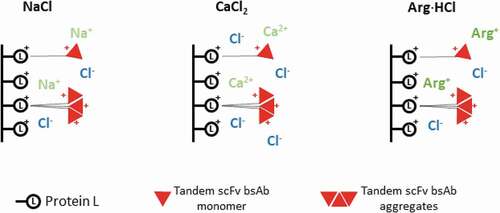
A commonly employed method to improve the antibody purity and yield in Protein A affinity chromatography is the use of salt additives in the elution buffer, which weakens the hydrophobic interaction between the antibody and Protein A ligand and reduces aggregation through chaotropic disruption, with the more chaotropic salts such as CaCl2 yielding products with enhanced purity compared to less chaotropic salts such as NaCl.Citation24 Arg·HCl has also been proposed to be an effective elution buffer additive for Protein A chromatography, as it favors the recovery of non-aggregated IgG due to its ability to suppress antibody aggregation and facilitate dissociation from Protein A through a salting-in mechanism.Citation25,Citation26 Furthermore, Arg·HCl has also been reported to be able to prevent aggregation and reduce viscosity at higher protein concentration, aid in protein refolding, and has been shown to inactivate enveloped viruses.Citation27–Citation30 In comparison to Protein A chromatography, the salt additive effect in Protein L affinity chromatography is less well understood. Until most recently, NaCl has been reported to be able to separate aggregated species in Protein L chromatography based on their different binding valencies,Citation31 suggesting the presence of a different mechanism from the previously reported chaotropic effect exerted by salt additives in the elution buffer of affinity chromatography.
Here, using blinatumomab biosimilar as a model tandem scFv bsAb molecule, we probed a range of different salt concentrations as well as salt types, including NaCl, CaCl2, and Arg·HCl, to reveal a preferential strengthening effect of the high molecular weight (HMW)-protein interaction compared to monomer-Protein L interaction. This resulted in an enhanced monomer-HMW separation that is governed by an interplay between electrostatic interactions, as well as other interactions such as hydrophobicity, cation-π interaction and hydrogen bonding, with Arg·HCl emerging as the most effective salt additive to yield the optimal product purity and recovery. Within a two-step purification process using Protein L affinity resin as the capture step, we demonstrate the purification of a final tandem scFv bsAb product comprising <1% HMW species and <100 ppm host cell proteins (HCP) with an overall yield of 65%.
Results
Preliminary evaluation of Protein L chromatography as capture step for tandem scFv bsAb molecules
We first set out to evaluate the performance of Protein L affinity chromatography as a capture step for tandem scFv bsAb molecules, by using blinatumomab biosimilar as a model tandem scFv bsAb molecule and TOYOPEARLTM AF-rProtein L-650F as a model Protein L affinity resin. TOYOPEARLTM AF-rProtein L-650F was chosen as the model Protein L affinity resin because, according to the product brochure, it has the highest binding capacity available on the market. The resin comprises a rigid polymethacrylate matrix combined with a recombinant ligand derived from the B4 domain of native Protein L.
As the recommended elution pH in the product brochure for antibody fragments is pH 2.0–3.0, we decided to use pH 3.0 (100 mM acetate) for the step elution as previously reported.Citation20 Under this elution condition, we observed that the low molecular weight (LMW) species were significantly reduced, from 26.6% in the cell culture supernatant (CCS) to 2.6% in the Protein L eluate, whereas the overall HMW species content was only slightly reduced, from 52.3% to 44.3%, yielding a monomer purity of 53.0% (). Similar results were also obtained using CaptoTM L and KANEKA KanCapTM L (data not shown).
Addition of salt additives in elution buffer for enhanced monomer-HMW species separation
In order to improve the separation of monomers from the HMW species, we screened three different salts, namely NaCl, CaCl2, and Arg·HCl, at varying concentrations as additives in the elution buffer. Using 50 mM, 70 mM, 100 mM, and 500 mM of the different salts in acetate buffer at pH 3.0, we were able to tune the amount of HMW species and monomer that eluted by a two-step elution process in the presence (peak 1) and absence (peak 2) of the salt additives, as demonstrated in the representative AKTA chromatogram and their corresponding HPLC-SEC chromatograms (, )). An increase in the concentration of salt additives led to an increase in monomer proportion that eluted in the presence of salt additives in peak 1, with a complete suppression, within detection limits, of the elution of the tandem scFv bsAb at 500 mM of salt additives. An analysis of the total peak area of peaks 1 and 2 showed that increasing concentrations of Arg·HCl resulted in a limited recovery of the tandem scFv bsAb at pH 3.0, especially beyond 100 mM Arg·HCl, with a similar trend observed for NaCl and CaCl2 albeit to a smaller extent (Supp Figure 1a-d). The total peak area, however, remains relatively constant when the peak obtained during cleaning in place (CIP) in the presence of 100 mM sodium hydroxide (NaOH) is taken into consideration (Supp Figure 1e-f). Taken together, this result may be attributed to the overly enhanced hydrophobic interaction of the tandem scFv bsAb at high salt concentrations, as has been reported for the purification of monoclonal antibodies by Protein A chromatography.Citation32
Figure 3. (a) Representative Protein L chromatogram illustrating the separation of the monomer and HMW species peaks in the presence of NaCl, CaCl2, or Arg·HCl at pH 3.0. Shown here is the elution obtained in the presence and absence of 100 mM Arg·HCl, pH 3.0. (b) HPLC-SEC chromatograms of the first (top) and second peak (bottom) corresponding to predominantly monomers and HMW species, respectively. (c) Purity profile of peaks 1 and 2 illustrating the proportion of HMW species and monomer that eluted in the presence of 50 mM, 70 mM, 100 mM, and 500 mM of NaCl, CaCl2, and L-arginine, along with the corresponding monomer recovery. (d) Purity profile of peaks 1 and 2 of eluates obtained in the presence of 50–500 mM Arg·HCl, along with the corresponding monomer recovery
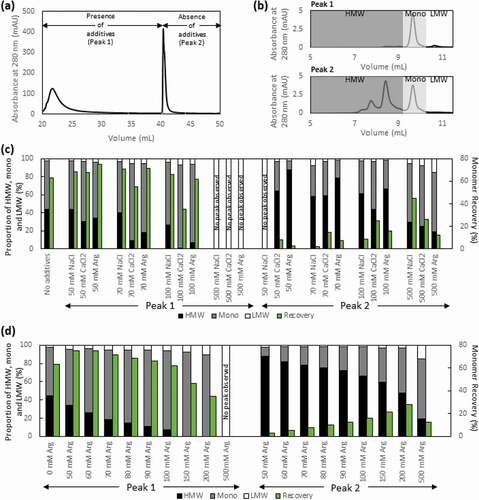
While CaCl2 yielded the lowest proportion of HMW species present in peak 1 across all the different concentrations ()), it also suppressed the elution of monomer, resulting in the subsequent co-elution of the monomer with HMW species in peak 2 in the absence of CaCl2. The eluate obtained in the presence of Arg·HCl contained slightly higher HMW species compared to that of CaCl2, but with lower proportions of monomer co-eluting with the HMW species in peak 2 in the concentrations screened. Hence, considering the balance between the purity and the distribution of monomer between peak 1 and 2, Arg·HCl stands out as the best salt additive ()). In order to further ascertain the optimal Arg·HCl concentration to be used in the elution buffer, a series of different Arg·HCl concentrations between 50 and 500 mM was screened. An increase in Arg·HCl concentration resulted in predominantly monomers in peak 1 and HMW species in peak 2, with 100 mM of Arg·HCl giving the optimal results in terms of monomer recovery and lowest HMW content ()).
Investigation of the mechanism involved in enhancing the monomer-HMW species separation in the presence of salt additives
To gain insight into the mechanism governing the enhanced monomer-HMW species separation in the presence of salt additives in the elution buffer, we performed a comparison of the elution peak 1 at different salt concentrations and salt types (–c)). We observed that, compared to the control experiment where no salt was added, broadening of the elution peak, along with an increase in retention time, was observed in the presence of salt additives, with the peak broadening and retention time delay increasing with increasing concentration of salt additives. This clearly indicates an overall strengthening of the interaction between tandem scFv bsAb and Protein L in the presence of these salt additives. In addition, the fact that both the monomeric and HMW species co-eluted at pH 3.0 in the absence of additives while mainly the monomer is eluted in the presence of additives suggests that the enhanced HMW-monomer separation obtained in the presence of these additives is achieved through the preferential strengthening of HMW-Protein L interaction, as compared to that of monomer-Protein L interaction.
Figure 4. The elution peak obtained in the presence of varying concentrations of Arg·HCl (a), NaCl (b) and CaCl2 (c) demonstrates the differential increase in retention time and peak broadening compared to the elution peak obtained in the absence of salt additives (green). (d) The conductivity of eluate peak 1 is plotted as a function of monomer (%) for all three salt additives. (e) The hydrophobicity of the solvent environment at different Arg·HCl concentrations was probed with nile red fluorescent dye
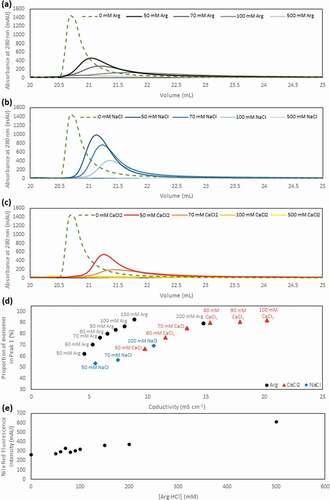
We next probed the relationship of monomer elution as a function of conductivity for each salt type and observed that an increase in conductivity resulted in an increase in the proportion of monomer that eluted in the presence of all three salt additives ()), as previously reported for NaCl.Citation31 This can be rationalized by the electrostatic interactions between Protein L and the tandem scFv bsAb. As both Protein L and the tandem scFv bsAb are positively charged at pH 3.0, increased salt concentrations will mask these repulsive interactions, therefore strengthening the tandem scFv bsAb-Protein L interactions. Due to their valency differences, the HMW-Protein L interaction will be preferentially strengthened compared to monomer-Protein L interaction, leading to enhanced monomer-HMW species separation.
Nevertheless, while similar conductivities were required for NaCl and CaCl2 to achieve similar amounts of monomer in the eluate, a lower conductivity yielded similar results in the presence of Arg·HCl ()). This suggests that, in addition to conductivity, other factors may also play a key role in the modulation of the Protein L binding avidity. Arginine is known to be able to form hydrogen bonds and cation-π interactions with proteinsCitation33,Citation34 and increases the hydrophobicity of the solvent environment.Citation35 We assessed the extent of hydrophobicity conferred by Arg·HCl at the concentrations used in this study by using the widely used phenoxazone-based nile red dye, which is known to be sensitive to the hydrophobicity of the environment.Citation36,Citation37 The increase in fluorescence intensity of the nile red dye with increasing concentration of Arg·HCl indicates an increase in hydrophobicity in the solvent environment at higher Arg·HCl concentrations (), which weakens the hydrophobic interactions between Protein L and tandem scFv bsAb.
The effect of the interplay between electrostatic interactions and other interactions such as hydrophobicity, cation-π interaction and hydrogen bond formation is further supported by the trend observed in the increase in the retention time of the eluate peak in the presence of salt additives (). The presence of CaCl2 resulted in the largest increase in retention time compared to the same concentrations of NaCl and Arg·HCl, with Arg·HCl showing the smallest increase in retention time. This can be explained by the greater masking of the repulsive positive charge interactions between Protein L and tandem scFv bsAb by CaCl2, which has twice the amount of chloride ions compared to NaCl (). On the other hand, while the increase in concentration of Arg·HCl also strengthens the Protein L-tandem scFv bsAb ionic interaction, its increased hydrophobicity and ability to form cation-π interaction and hydrogen bonds results in a slight weakening of this interaction, therefore resulting in the overall smallest increase in retention time ().
Development of a two-step process for the purification of tandem scFv bsAB
In order to develop a full process for the purification of tandem scFv bsAb, we performed a design of experiment (DoE) using different concentrations of Arg·HCl, NaCl, and CaCl2 at varying pH to optimize an effective wash buffer for the removal of HCP and other contaminants, as these salts have been reported to be effective wash buffers for Protein A chromatography.Citation38,Citation39 The optimal wash condition obtained in this manner based on the DoE Unicorn 6.3 evaluation software is 513 mM Arg·HCl, 87 mM NaCl at pH 6.9 (Supp Table 1, Supp Figure 2), suggesting that Arg·HCl has the most significant effect on both HCP removal and product recovery, whereas CaCl2 exerts a negligible effect. The enhanced HCP removal in the presence of Arg·HCl is likely to occur via the weakening of nonspecific hydrophobic interactions, as has been proposed in previous studies for Protein A chromatography.Citation38,Citation39 The response surface plots of Arg·HCl and NaCl at different pH values reveal that lower HCP content was obtained at the higher Arg·HCl concentrations and lower NaCl concentrations, whereas higher recovery was obtained at lower Arg·HCl and NaCl concentrations, with low pH values yielding the lowest HCP removal and overall product recovery (Supp Figure 2). Based on the predicted minimum HCP removal, the DoE optimized wash buffer gives an HCP fold reduction of 5481 times, which is a 3-fold improvement from the HCP fold reduction of 1854 times when equilibration (EQ) buffer is used; the minimum predicted recovery is very similar in both cases, yielding 75.7% and 77.6% for the DoE optimized wash buffer and EQ wash buffer, respectively (Supp Table 2).
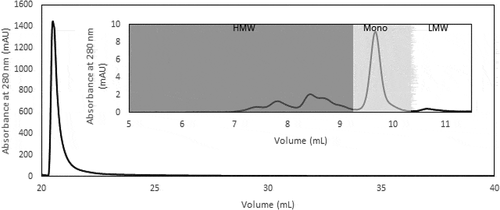
To validate the performance of the overall Protein L chromatography protocol, CCS was loaded onto the equilibrated Protein L column at 10 mg monomer/mL resin. Five column volumes (CVs) of the optimized wash buffer was subsequently used, followed by 5 CVs of 50 mM HEPES buffer, pH 7.4, so as to reduce the conductivity of the resultant eluate, which was obtained by elution in 100 mM Arg·HCl, pH 3.0 acetate buffer. The eluate obtained in this manner yielded a recovery of 81.3%, an HCP fold reduction of about 3000 times, and a reduction of HMW species from 66.5% in the CCS to 7.1% ().
Table 1. Verification run for ToyopearlTM AF-rProtein L-650F, where 10 mg of tandem scFv bsAb monomer containing CCS was loaded per mL of resin, with 5 CVs each of DoE optimized wash buffer followed by 50 mM HEPES pH 7.4 buffer and finally eluting with 100 mM Arg·HCl acetate buffer at pH 3.0. Optimization of CaptoTM adhere conditions were performed in bind and elute mode
Finally, we performed a polishing step using CaptoTM adhere multimodal resin to investigate the quality of the product after a single polishing step. Here, three different salt concentrations, 300 mM, 350 mM, 400 mM NaCl in 50 mM MES pH 6.0 buffer were screened as the elution buffer in the bind and elute mode. The results show that less than 400 mM NaCl was required to give HCP content less than 100 ppm and less than 1% HMW species in the eluate. This, however, comes with a compromise of the overall recovery, with lower salt concentrations yielding lower overall recovery. Hence, 350 mM NaCl appears to be the optimal condition at 50 mM MES, pH 6.0 buffer ().
The SEC profiles of CCS, Protein L eluate and CaptoTM adhere eluate show the progressive removal of the HMW species and HCP with each purification step, highlighting the relatively pure product obtained after Protein L chromatography, with small amounts of HMW species present, and a highly pure product obtained after a single polishing step ()). This is corroborated by the SDS-PAGE gel, which indicates high purity products obtained after both capture and polishing steps, with negligible amounts of impurities present in the sample, within the detection limits of the gel ()). Through a two-step purification process (Supp Figure 3), a pure tandem scFv bsAb product with HCP content less than 100 ppm and less than 1% HMW species can be achieved.
Discussion
Tandem scFv bsAbs represent one of the promising formats of recombinant bsAbs that have yielded exemplary clinical results, and many are currently in clinical development.Citation4–Citation9 It is therefore of the utmost relevance to develop downstream purification processes that allow for the efficient and effective isolation of these types of antibodies.
It has been reported that antibodies lacking the Fc region have an increased aggregation propensity compared to their parental monoclonal antibodies,Citation11,Citation12 and in this study, we similarly observed that the CCS for blinatumomab biosimilar contained high amounts of aggregates, ranging between 52.3% and 66.5%. While a step elution with pH 3.0 acetate buffer reduced the amount of aggregates by 8.0%, we demonstrated here that the amount of aggregates can be most effectively reduced by more than 40% using 100 mM Arg·HCl salt additive in pH 3.0 acetate buffer in a single-step Protein L chromatography.
As previously reported for Protein A chromatography,Citation24–Citation26 careful alteration of the interactions between the affinity ligand and the target molecule through the addition of salt additives can result in enhanced purity of the eluate. Interestingly, we observed that this occurs via a mechanism that preferentially strengthens the HMW-Protein L interaction as compared to that of monomer-Protein L interaction, rather than the widely reported chaotropic effect exerted by salt additives observed in Protein A chromatography, which dissociates protein-protein complexes.Citation24–Citation26 An enhanced chaotropic effect induced by salt additives in the elution buffer should ease the elution of bound proteins, especially for the monomer, with more efficient dissociation obtained with increasing salt concentrations and with the stronger chaotrope CaCl2 compared to NaCl. However, these results were not observed in our study. On the contrary, in our study, compared to control experiment, the eluted monomer peak of tandem scFv bsAb was broadened and delayed in the presence of salt additives in elution buffer (), and this strengthening effect increased with the concentration of salt additives. This preferential strengthening effect is different from the widely reported chaotropic effect of salt additives in elution buffer for Protein A chromatography,Citation24–Citation26 suggesting the presence of a different dominant interaction between tandem scFv bsAb and Protein L compared to that of antibody and Protein A.
A recent studyCitation31 reported the use of controlled conductivity at low pH, as demonstrated by varying NaCl concentrations, in Protein L chromatography to separate different antibody formats by their binding valency. Our results corroborate well with their finding that increased conductivity leads to better monomer-aggregate separation. Furthermore, we observed that lower conductivities were required for Arg·HCl to achieve similar results as compared to NaCl and CaCl2, suggesting that other properties of the salts, such as their hydrophobicity, cation-π, and hydrogen bonding propensity, can also affect the interaction between Protein L and the monomers or HMW species, as supported by the differential increase in retention time of the eluate in the presence of the different salt additives. As Arg·HCl contains a protonated guanidinium group that can form ionic and hydrogen bonds, a future study might include the use of guanidine HCl to mechanistically separate the effect of the arginine peptide backbone from the guanidinium ion. In addition, we find that there is a threshold of salt concentration, above which the overall recovery of the product will be severely compromised. This may be attributed to the overly enhanced hydrophobic interaction of both HMW and monomeric species at high salt concentrations, as has been reported for the purification of monoclonal antibodies by Protein A chromatography.Citation32 Taken together, this suggests that hydrophobicity, cation-pi interaction and hydrogen bonding, along with conductivity, play crucial roles in modulating the Protein L binding avidity and enabling the effective separation of the monomer from aggregates.
Using Protein L chromatography, previous studies have reported the purification of a bovine tandem scFv bsAb, achieving an overall purity of 50 – 70% using a step elution of 100 mM glycine at pH 3.0Citation20 and requiring the need to perform an additional Protein A or Protein G column in flow-through mode and SEC when performing the purification in large scale for enhanced purity.Citation19 Here, after a one-step Protein L affinity chromatography purification of Chinese hamster ovary (CHO) cell-derived tandem scFv bsAb, we show that it is possible to achieve more than 90% purity using a step elution of 100 mM Arg·HCl in acetate buffer pH 3.0.
In addition to investigating the ability of Protein L chromatography to remove aggregates while maintaining high recovery, we also probed the use of Protein L chromatography to remove HCP and found that Arg·HCl in the presence of minimal amounts of other salts was the most effective at HCP removal. Interestingly, while previous studies for Protein A chromatography report more favorable HCP removal when higher concentrations of NaCl were used in combination with Arg·HCl,Citation39 in our study we observed that lower concentrations of NaCl in the presence of Arg·HCl was more effective at removing HCP, suggesting that ionic interactions between charged contaminants bound to charged residues of the affinity chromatography matrix, including the resin, affinity ligand, and/or target bound to the ligand, may play a smaller role in our case.
In conclusion, through the investigation of the use of three different salt additives, namely NaCl, CaCl2, and Arg·HCl, in the elution buffer for the purification of tandem scFv bsAb using Protein L affinity chromatography, we propose a different mechanism for the enhanced HMW-monomer separation observed in the presence of the salt additives. Arg·HCl, in particular, was the choice of salt additive due to the optimal preferential strengthening of the HMW-Protein L interaction, compared to that of monomer-Protein L, governed by the best balance of electrostatic interactions, as well as other interactions such as hydrophobicity, cation-π interaction, and hydrogen bonding, which resulted in the optimal recovery and purity at 100 mM Arg·HCl. As a proof-of-concept of the feasibility of using Protein L affinity chromatography for the purification of tandem scFv bsAb, a two-step purification process was developed. The overall HMW species was reduced by 59% and HCP content by 3000 fold, giving a monomeric step-yield of 81% after the capture step. Using CaptoTM adhere as the next polishing step, HMW species and HCP can be reduced to <1% and <100 ppm, respectively, with overall yield of 65%, demonstrating the potential of using Protein L affinity chromatography for the purification of aggregation-prone antibodies without Fc regions and/or containing appropriate kappa light chain regions within a two-step process.
Materials and methods
All buffers, salts, and reagents were obtained from Merck Millipore, except L-Arginine mono-hydrochloride, which was obtained from Sigma-Aldrich.
Blinatumomab biosimilar culture
The bispecific blinatumomab biosimilar targeting CD19 and CD3 was produced in a recombinant CHO K1 clone generated using a bicistronic vector and stable transfection. The bispecific vector expresses the gene encoding blinatumomab and a zeocin resistant gene in one transcript through the use of internal ribosome entry element (IRES). The amino acid sequence of blinatumomab was obtained from the publicly available International ImMunoGeneTics Information System (IMGT). Production was operated using fed-batch mode in 5 L glass stirred tank bioreactors using protein-free media. The cells were harvested by centrifugation at 4000 × g for 20 min at room temperature. After centrifugation, the supernatant was filtered through 0.22 µm membrane. The filtrate was stored in sterile containers until further processing.
ÄKTATM chromatography
Unless otherwise stated, all chromatographic runs were performed at 2 min residence time and all optimization runs were performed with a load of 1 mg/mL of monomeric blinatumomab biosimilar, with the exception of the final Protein L verification run, which was performed with 10 mg/mL of monomeric blinatumomab biosimilar. All chromatography media were packed in 1 mL TricornTM series columns with a bed height of 5.1 cm (GE Healthcare), with experiments conducted on an ÄKTATM Avant 25 (GE Healthcare).
All columns containing ToyopearlTM AF-rProtein L-650F (Tosoh Bioscience) were equilibrated with 50 mM HEPES, 120 mM NaCl pH 7.4, before loading the appropriate amount of CCS. For the screening of additives for enhanced aggregate-monomer separation, the column was washed with 10 CVs of equilibration buffer after loading the appropriate amount of CCS before proceeding with elution using 100 mM acetate pH 3.0 containing varying concentrations of NaCl, CaCl2, or Arg·HCl as required, followed by eluting using 100 mM acetate pH 3.0 in the absence of salt additives, collecting the eluate peak from 5 mAu – 5 mAu. The verification run was performed using the optimized conditions, which comprises 513 mM Arg·HCl, 87 mM NaCl, pH 6.7 as wash buffer (5 CV), followed by 50 mM HEPES, pH 7.4 (5 CV) to reduce the conductivity, and finally eluting with 100 mM Arg·HCl, acetate buffer at pH 3.0.
The polishing step was performed using CaptoTM adhere resin (GE Healthcare) in bind and elute mode. The column was equilibrated in 50 mM MES, pH 6.0 buffer. The eluate obtained from Protein L chromatography was adjusted to pH 6.0 and diluted 3 times to a conductivity of 6–7 mS/cm before loading onto the column, followed by elution in 50 mM MES, pH 6.0 buffer containing 300 mM, 350 mM, or 400 mM NaCl, collecting the eluate peak from 5 mAu – 5 mAu.
Antibody concentration and purity analysis
The antibody concentration and purity were analyzed by HPLC-SEC using a TSKgel G3000SWXL column (7.8 mm i.d. x 30 cm; Tosoh Bioscience) at a flow rate of 0.6 mL/min. The mobile phase consisted of 0.2 M L-arginine, 0.05 M MES, 5 mM EDTA, 0.05% sodium azide (w/w), pH 6.5. The UV absorbance was monitored at 280 nm, with the resultant concentrations determined based on the area under the peaks as compared to a calibration curve obtained using standard samples. The amount of aggregates and fragments present in the sample was calculated using the area of peaks that eluted before and after the monomeric peak, respectively. SDS-PAGE, along with staining with InstantBlueTM, was also used to investigate the purity of the sample.
Residual host cell protein and DNA analysis
The CHO HCP content was determined by the microtiter plate enzyme-linked immunosorbent assay (ELISA) 3rd Generation CHO HCP kit (Cygnus Technologies), according to the manufacturer’s instructions. Data acquisition was performed with the Gen5TM software on a SynergyTM 2 plate reader (BioTek).
The CHO DNA content was measured using a QX200TM Droplet DigitalTM PCR System (Bio-Rad Laboratories), according to the manufacturer’s instructions. Briefly, samples were digested with proteinase K (0.2 mg/mL in 0.5% SDS, 16 h, 50°C), followed by inactivation (10 min, 95°C). DNA extraction from the resultant solution was subsequently performed using the QIAamp viral RNA mini kit (QIAGEN). As described on QIAamp® Viral RNA Mini Kit Handbook page 10, this kit can purify both RNA and DNA in parallel.Citation40 The protocol used for the DNA extraction here is that recommended for the purification of cellular, bacterial, or viral DNA from the handbook page 27 and page 36.Citation40 The method has also been reported to be used for CHO host cell DNA extraction.Citation41 The PCR reaction mixture was prepared by adding ddPCRTM supermix for residual DNA quantification (Bio-Rad Laboratories), ddPCRTM CHO residual DNA quantification assay (Bio-Rad Laboratories), XenoTM VICTM primer probe mix (Applied Biosystems), XenoTM DNA control (Applied Biosystems) and the extracted DNA sample together. The resultant reaction mixture (20 µL) and the droplet generation oil (70 µL) were loaded into an eight-channel disposable droplet generator cartridge for the generation of droplets. The generated droplets were then gently transferred into a 96-well PCR plate and heat-sealed with PX1TM PCR plate sealer (Bio-Rad Laboratories). The PCR reaction (10 min at 95°C; 40 cycles of 30 s at 94°C followed by 1 min at 60°C; 10 min at 98°C) was performed with C1000 TouchTM thermal cycler (Bio-Rad Laboratories). Data recording and analysis were performed with QuantaSoft analysis software (Bio-Rad Laboratories). The conversion of DNA copy number to DNA concentration was based on CHO host cell DNA standards (Applied Biosystems).
Hydrophobicity measurement by nile red
Nile red stock solutions were prepared by dissolving nile red powder into dimethyl sulfoxide and then diluted into water and used at a final concentration of 0.5 µM. The hydrophobicity of the different concentrations of Arg·HCl was quantified by monitoring the fluorescence emissions in a black, non-binding 96-well microplate (Greiner Bio-one) using an Infinite M200 Pro plate reader (Tecan). Excitation wavelength was set at 532 ± 10 nm and the fluorescence emission was collected at 654 ± 20 nm with a gain setting of 100.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Word (205 KB)Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Gokbuget N, Dombret H, Bonifacio M, Reichle A, Graux C, Faul C, Diedrich H, Topp MS, Bruggemann M, Horst HA, et al. Blinatumomab for minimal residual disease in adults with B-precursor acute lymphoblastic leukemia. Blood. 2018;131(14):1522–10. doi:https://doi.org/10.1182/blood-2017-08-798322.
- Kantarjian H, Stein A, Gokbuget N, Fielding AK, Schuh AC, Ribera JM, Wei A, Dombret H, Foa R, Bassan R, et al. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N Engl J Med. 2017;376(9):836–47. doi:https://doi.org/10.1056/NEJMoa1609783.
- Oldenburg J, Mahlangu JN, Kim B, Schmitt C, Callaghan MU, Young G, Santagostino E, Kruse-Jarres R, Negrier C, Kessler C, et al. Emicizumab prophylaxis in hemophilia A with inhibitors. N Engl J Med. 2017;377(9):809–18. doi:https://doi.org/10.1056/NEJMoa1703068.
- Labrijn AF, Janmaat ML, Reichert JM, Parren PWHI. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18(8):585–608. doi:https://doi.org/10.1038/s41573-019-0028-1.
- Kontermann RE. Dual targeting strategies with bispecific antibodies. MAbs. 2012;4(2):182–97. doi:https://doi.org/10.4161/mabs.4.2.19000.
- Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs. 2017;9(2):182–212. doi:https://doi.org/10.1080/19420862.2016.1268307.
- Chames P, Baty D. Bispecific antibodies for cancer therapy: the light at the end of the tunnel? MAbs. 2009;1(6):539–47. doi:https://doi.org/10.4161/mabs.1.6.10015.
- Kontermann RE. Recombinant bispecific antibodies for cancer therapy. Acta Pharmacol Sin. 2005;26(1):1–9. doi:https://doi.org/10.1111/aphs.2005.26.issue-1.
- Baeuerle PA, Reinhardt C. Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res. 2009;69(12):4941–44. doi:https://doi.org/10.1158/0008-5472.CAN-09-0547.
- Sasso EH, Silverman GJ, Mannik M. Human IgA and IgG F(ab’)2 that bind to staphylococcal protein A belong to the VHIII subgroup. J Immunol. 1991;147:1877–83.
- Garber K. Bispecific antibodies rise again. Nat Rev Drug Discov. 2014;13(11):799–801. doi:https://doi.org/10.1038/nrd4478.
- Taki S, Kamada H, Inoue M, Nagano K, Mukai Y, Higashisaka K, Yoshioka Y, Tsutsumi Y, Tsunoda S. A novel bispecific antibody against human CD3 and ephrin receptor A10 for breast cancer therapy. PLoS One. 2015;10(12):e0144712. doi:https://doi.org/10.1371/journal.pone.0144712.
- Dorken B, Riethmuller G, Kufer P, Lutterbuse R, Bargou R, Loffler A. CD19XCD3 specific polypeptides and uses thereof. United States patent US 7,575,923 B2. 2009.
- Mack M, Riethmuller G, Kufer P. A small bispecific antibody construct expressed as a functional single-chain molecule with high tumor cell cytotoxicity. Proc Natl Acad Sci USA. 1995;92(15):7021–25. doi:https://doi.org/10.1073/pnas.92.15.7021.
- Loffler A, Kufer P, Lutterbuse R, Zettl F, Daniel PT, Schwenkenbecher JM, Riethmuller G, Dorken B, Bargou RC. A recombinant bispecific single-chain antibody, CD19 X CD3, induces rapid and high lymphoma-directed cytotoxicity by unstimulated T lymphocytes. Blood. 2000;95(6):2098–103. doi:https://doi.org/10.1182/blood.V95.6.2098.
- Brischwein K, Schlereth B, Guller B, Steiger C, Wolf A, Lutterbuese R, Offner S, Locher M, Urbig T, Raum T, et al. MT110: A novel bispecific single-chain antibody construct with high efficacy in eradicating established tumors. Mol Immunol. 2006;43(8):1129–43. doi:https://doi.org/10.1016/j.molimm.2005.07.034.
- Pendzialek J, Roose K, Smet A, Schepens B, Kufer P, Raum T, Baeuerle PA, Muenz M, Saelens X, Fiers W. Bispecific T cell engaging antibody constructs targeting a universally conserved part of the viral M2 ectodomain cure and prevent influenza A virus infection. Antiviral Res. 2017;141:155–64. doi:https://doi.org/10.1016/j.antiviral.2017.02.016.
- Kufer P, Lutterbuse R, Kohleisen B, Zeman S, Bauerle P. Pharmaceutical compositions comprising bispecific anti-CD3, anti-CD-19 antibody constructs for the treatment of B-cell related disorders. United States patent US 7,635,472 B2. 2009.
- Spiesberger K, Paulfranz F, Egger A, Reiser J, Vogl C, Rudolf-Scholik J, Mayrhofer C, Grosse-Hovest L, Brem G. Large-scale purification of r28M: A bispecific scFv antibody targeting human melanoma produced in transgenic cattle. PLoS One. 2015;10(10):e0140471. doi:https://doi.org/10.1371/journal.pone.0140471.
- Grosse-Hovest L, Muller S, Minoia R, Wolf E, Zakhartchenko V, Wenigerkind H, Lassnig C, Besenfelder U, Muller M, Lytton SD, et al. Cloned transgenic farm animals produce a bispecific antibody for T cell-mediated tumor cell killing. Proc Natl Acad Sci USA. 2004;101(18):6858–63. doi:https://doi.org/10.1073/pnas.0308487101.
- Bjorck L, Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol. 1988;140:1194–97.
- Graille M, Stura EA, Housden NG, Beckingham JA, Bottomley SP, Beale D, Taussig MJ, Sutto BJ, Gore MG, Charbonnier JB. Complex between peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure. 2001;9(8):679–87. doi:https://doi.org/10.1016/S0969-2126(01)00630-X.
- Nilson BH, Logdberg L, Kastern W, Bjorck L, Akerstrom B. Purification of antibodies using protein L-binding framework structures in the light chain variable domain. J Immunol Methods. 1993;164(1):33–40. doi:https://doi.org/10.1016/0022-1759(93)90273-A.
- Tustian AD, Endicott C, Adams B, Mattila J, Bak H. Development of purification processes for fully human bispecific antibodies based upon modification of protein A binding avidity. MAbs. 2016;8(4):828–38. doi:https://doi.org/10.1080/19420862.2016.1160192.
- Arakawa T, Philo JS, Tsumoto K, Yumioka R, Ejima D. Elution of antibodies from a Protein-A column by aqueous arginine solutions. Protein Expr Purif. 2004;36(2):244–48. doi:https://doi.org/10.1016/j.pep.2004.04.009.
- Ejima D, Yumioka R, Tsumoto K, Arakawa T. Effective elution of antibodies by arginine and arginine derivatives in affinity column chromatography. Anal Biochem. 2005;345(2):250–57. doi:https://doi.org/10.1016/j.ab.2005.07.004.
- Inoue N, Takai E, Arakawa T, Shiraki K. Specific decrease in solution viscosity of antibodies by arginine for therapeutic formulations. Mol Pharmaceutics. 2014;11(6):1889–96. doi:https://doi.org/10.1021/mp5000218.
- Tsumoto K, Umetsu M, Kumagai I, Ejima D, Philo JS, Arakawa T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol Prog. 2004;20(5):1301–08. doi:https://doi.org/10.1021/bp0498793.
- Chen J, Liu Y, Wang Y, Ding H, Su Z. Different effects of L-arginine on protein refolding: suppressing aggregates of hydrophobic interaction, not covalent binding. Biotechnol Prog. 2008;24(6):1365–72. doi:https://doi.org/10.1002/btpr.93.
- Meingast C, Heldt CL. Arginine-enveloped virus inactivation and potential mechanisms. Biotechnol Prog. 2019;e2931.
- Chen C, Wakabayashi T, Muraoka M, Feng S, Chia WS, Chong CK, Ching TJ, Soehano I, Shimizu Y, Igawa T, et al. Controlled conductivity at low pH in Protein L chromatography enables separation of bispecific and other antibody formats by their binding valency. MAbs. 2019;11(4):632–38. doi:https://doi.org/10.1080/19420862.2019.1583996.
- Bolton GR, Selvitelli KR, Iliescu I, Cecchini DJ. Inactivation of viruses using novel protein A wash buffers. Biotechnol Prog. 2015;31(2):406–13. doi:https://doi.org/10.1002/btpr.2024.
- Shukla D, Trout BL. Interaction of arginine with proteins and the mechanism by which it inhibits aggregation. J Phys Chem B. 2010;114(42):13426–38. doi:https://doi.org/10.1021/jp108399g.
- Shukla D, Zamolo L, Cavallotti C, Trout BL. Understanding the role of arginine as an eluent in affinity chromatography via molecular computations. J Phys Chem B. 2011;115(11):2645–54. doi:https://doi.org/10.1021/jp111156z.
- Das U, Hariprasad G, Ethayathulla AS, Manral P, Das TK, Pasha S, Mann A, Ganguli M, Verma AK, Bhat R, et al. Inhibition of protein aggregation: supremolecular assemblies of arginine hold the key. PLoS One. 2007;2(11):e1176. doi:https://doi.org/10.1371/journal.pone.0001176.
- Sackett DL, Wolff J. Nile red as a polarity-sensitive fluorescent probe of hydrophobic protein surfaces. Anal Biochem. 1987;167(2):228–34. doi:https://doi.org/10.1016/0003-2697(87)90157-6.
- Fowler SD, Greenspan P. Application of nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem. 1985;33(8):833–36. doi:https://doi.org/10.1177/33.8.4020099.
- Sun S, Gallo C. Arginine derivative wash in protein purification using affinity chromatography. United States patent US 7,714,111 B2. 2010.
- Frauenschuh A, Bill K. Wash solution comprising arginine for the affinity chromatography step in antibody purification. United States patent US 2017/0,044,211 A1. 2017.
- QIAamp® Viral RNA. Mini handbook; March 2018. version. QIAGEN.
- Nian R, Zhang W, Tan L, Lee J, Bi X, Yang Y, Gan HT, Gagnon P. Advance chromatin extraction improves capture performance of protein A affinity chromatography. J Chromatogr A. 2016;1431:1–7. doi:https://doi.org/10.1016/j.chroma.2015.12.044.