ABSTRACT
The aim of this study was to investigate the antibiotic resistance profiles and staphylococcal enterotoxins/enterotoxin-likes genes (se/sel) of Staphylococcus aureus from food. About 93 S. aureus strains were screened and 43 strains harbored se/sel genes, which were from raw meat, pickles, cooked meat, egg products, bean products, aquatic products, grains and fresh vegetables, respectively. Among the 21 se/sel genes, selx gene was found in 30/93 (32.26%) strains, followed by seb, ser, sea, sec, selp, sell and set. About 50/93 strains carried 10 antibiotic resistance genes. The prevalence rate of norA, chlA and grlA genes was relatively high, followed by ermC, tetA, aac6ʹ/aph2”, ermB, tetM, msrA and blaZ. Sixty-two strains of the 93 S. aureus isolates (62/93, 66.67%) were multidrug-resistant (≥3 antimicrobial agents).
RESUMEN
El presente estudio se orientó a investigar los perfiles de resistencia a los antibióticos y los genes de enterotoxinas estafilocócicas y de aquellos parecidos a enterotoxinas (se/sel) de S. aureus aislada de alimentos. Con este objetivo se cribaron 93 cepas de S. aureus, 43 de las cuales albergaban genes se/sel, procedentes de carne cruda, pepinillos, carne cocida, productos de huevo, productos de frijol, productos acuáticos, granos y verduras frescas. En 30 de las 93 cepas (32,26%) se encontró el gen selx entre los 21 genes se/sel y, en orden descendente, los genes seb, ser, sea, sec, selp, sell y set. Asimismo, se constató que 50 de las 93 cepas albergaban 10 genes de resistencia a los antibióticos. La tasa de prevalencia de los genes norA, chlA y grlA fue relativamente elevada siguiéndole, en orden descendente, las de los genes ermC, tetA, aac6ʹ/aph2”, ermB, tetM, msrA y blaZ. De las 93 cepas de S. aureus aisladas (62/93, 66,67%) 62 son resistentes a diversos medicamentos (≥tres agentes antimicrobianos).
Introduction
The seriousness of the problem of bacterial resistance is confirmed by the number of deaths associated with drug-resistant bacterial infections (Chudobova et al., Citation2014). Currently, antibiotic resistance is rising to dangerously high levels in all parts of the world (Tang et al., Citation2016). Resistant microorganisms are able to withstand an attack of antimicrobial medicines, so that standard treatments become ineffective and infections persist increasing the risk of spreading to others (WHO, Citation2014). With the discovery of multidrug-resistant strains in the broader community, public health officials have begun to realize the potential danger of the spread of these antibiotic-resistant bacteria in food sources (Chudobova et al., Citation2014).
Staphylococcus aureus (S. aureus) is an abundant bacterium occurring as commensal flora of humans and various animal species (Sung, Lloyd, & Lindsay, Citation2008). It is also a common bacterium found on the skin and nasal passages of healthy people (Yan et al., Citation2015). Approximately 25–40% of the population is colonized with S. aureus (Frana et al., Citation2013). Beyond asymptomatic carriage, S. aureus is associated with a wide variety of diseases in humans and animals, ranging from minor, uncomplicated skin and soft tissue infections, intramammary infection in dairy ruminants, food poisoning to more serious infections, such as pneumonia, bacteremia, meningitis, sepsis and pericarditis (Carfora et al., Citation2016; Jansen, Girgenti, Scully, & Anderson, Citation2013; Meemken et al., Citation2013; Monecke et al., Citation2011). In recent years, the increased frequency of methicillin-resistant S. aureus (MRSA) associated with nosocomial infections and their tendency to be multidrug-resistant, leading to reduced effectiveness of antibiotics and growing health-care costs, has made the pathogen a major public health concern (Jernigan et al., Citation1995; Kejela & Bacha, Citation2013; Xu et al., Citation2014).
The increasing resistance to antibiotics can be connected to antibiotic overuse and requires to be addressed promptly (Netsvyetayeva et al., Citation2014). The treatment of infections caused by S. aureus has been complicated by antimicrobial resistance in the bacteria (Jackson, Davis, & Barrett, Citation2013). For example, benzyl penicillin was no longer effective for treatment of most S. aureus infections after its introduction for use because of the acquisition of plasmid-encoded β-lactamase (Chambers, Citation2001; Sabath, Wheeler, Laverdiere, Blazevic, & Wilkinson, Citation1977). Penicillin-resistant S. aureus became pandemic throughout the late 1950s and early 1960s. Methicillin is β-lactam antibiotic invented to treat penicillin-resistant S. aureus; however, MRSA was reported 2 years after the antibiotic was introduced in 1961 (DeLeo & Chambers, Citation2009; Kejela & Bacha, Citation2013; Simonetti et al., Citation2011). It is estimated that 1.5% of the population (~4.1 million persons) is colonized with MRSA leading to at least 94,000 invasive infections and over 18,000 deaths annually in the United States (Frana et al., Citation2013). On the other hand, S. aureus possesses five major classical types of staphylococcal enterotoxins (SEs: SEA to SEE), and other non-classical SE-like toxins (SEls: SEG to SElX), which are mainly associated with food poisoning outbreaks (Puah, Chua, & Tan, Citation2016). Therefore, enterotoxigenic S. aureus with antibiotic resistance in food would pose a great threat to human health (Merz, Stephan, & Johler, Citation2016; Puah et al., Citation2016; Tang et al., Citation2015; Xing et al., Citation2016; Xu et al., Citation2014).
Due to the increasing risk of food sources S. aureus isolates for consumers, more investigation about the prevalence and characterization of food sources S. aureus strains should be executed. Therefore in this study, we examined the antimicrobial susceptibility patterns of S. aureus isolated from different food sources, and investigated their carriage of antibiotic resistance genes and staphylococcal enterotoxins/enterotoxin-likes (se/sel) genes.
Materials and methods
Isolation and identification of S. aureus
A total of 93 isolates were obtained from different food samples which were collected in supermarket, food market and retail stores in Chengdu City of Sichuan Province in the year 2014. All the isolates were identified as S. aureus species by biochemical tests, coagulase tests and S. aureus-specific polymerase chain reaction (PCR) assay.
Antimicrobial susceptibility testing
Susceptibility to 14 antimicrobial agents for all 93 isolates was performed by the disc diffusion procedure recommended by the Clinical and Laboratory Standards Inst. guidelines ([CLSI] Clinical and Laboratory Standards Inst, Citation2012) and other reported references (Jackson et al., Citation2013; Udegbunam, Udegbunam, & Anyanwu, Citation2014). Escherichia coli ATCC 25922 and S. aureus ATCC 25923 were used as a quality control strains.
DNA extraction
DNA extraction was conducted by using microwave oven heating method described by Wang et al. (Citation2015) in our lab. Briefly, all strains were grown in tryptic soy broth culture overnight at 37℃. Approximately, for each strain, 1 mL cell suspension was centrifuged at 12,000 r/min for 2 min, at 4℃, and the pellet was suspended in 1 mL Tris-HCl and EDTA (TE) buffer (10 mM Tris-HCl and 1 mM EDTA, pH=8.0), then 200 µL suspension was transferred into a new Eppendorf (EP) tube and centrifuged again, and the pellet was re-suspended in 100 µL TE buffer, treated in a microwave oven at rated power (800 W) for 60 s and centrifuged at 12,000 r/min for 2 min. The supernatant was transferred to a sterile tube and stored at −20℃ for PCR amplification.
Detection of antibiotic resistance gene and se/sel gene
The primer sequences of antibiotic resistance genes and se/sel genes are shown in and . Antibiotic resistance genes (blaTEM, oxa, mecA, blaZ, aac6ʹ/aph2”, tetM, tetA, ermA, ermB, ermC, msrA, mefA, grlA, norA, vanA, and chlA), staphylococcal enterotoxins genes (sea, seb, sec, sed, see, seg, seh, sei, ser, ses and set) and staphylococcal enterotoxin-likes genes (selj, selk, sell, selm, seln, selo, selp, selq, selu and selx) were detected by PCR amplification. About 20 μL reaction mixture for PCR assay was prepared as follows: 2 μL of 10×PCR buffer (Mg2+ free), 1.2 μL of deoxy-ribonucleoside triphosphate (dNTP) mixture (2.5 mM), 1.6 μL of 25 mM MgCl2, 0.5 μL of 20 μM corresponding primers, 1 μL of DNA sample, 0.2 μL of TaKaRa® Taq DNA polymerase (5 U/μL), up to 20 μL with H2O. PCR amplification conditions were as follows: initial denaturation at 95℃ for 5 min; 35 cycles of denaturation at 95℃ for 40 s, at annealing temperatures (seen in and ) for 50 s, and extension at 72℃ for 40 s, followed by a final extension at 72℃ for 10 min. The PCR products were visualized by Ultraviolet ray (UV) transillumination (Universal Hood II, Bio-Rad, Italy) after electrophoresis on 1.0% (w/v) agarose gels (Hydragene) with 0.5 mg/mL ethidium bromide (Sigma) in 0.5× Tris-Borate-EDTA (TBE, 0.89M Tris, 0.89M boric acid and 0.02M EDTA) buffer by a DYY-6C electrophoresis system (Beijing Liuyi Instrument Factory, Beijing). The positive PCR product for each gene was sequenced to ensure the reliability of amplification.
Table 1. the primer sequences of antibiotic resistance genes used in this study.
Tabla 1. Secuencias iniciadoras de los genes de resistencia a los antibióticos usados en este estudio.
Table 2. The primer sequences of se/sel genes used in this study.
Tabla 2. Secuencias iniciadoras de los genes se/sel usados en este estudio.
Table 3. The antibiotic agents and their concentrations used in this study.
Tabla 3. Agentes antibióticos y concentraciones usadas en este estudio.
Statistical analysis
Statistical analyses were performed using SPSS13.0 (SPSS, Chicago, IL, USA). Pearson’s and Yates’ chi-square tests were used to assess inter-group significance. Statistical significance was assumed at P < 0.05.
Results
The antimicrobial susceptibility pattern
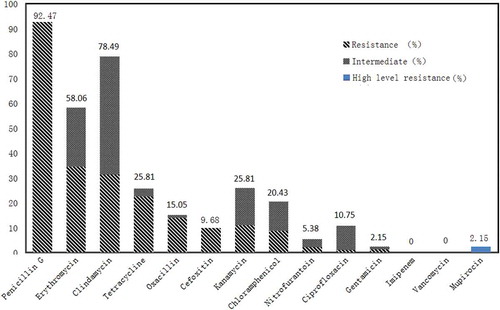
All 93 S. aureus food isolates had a diverse antimicrobial resistance profile (). Altogether, 49 kinds of antimicrobial resistance patterns are listed in . Among them, six isolates were resistant to one drug, followed by 25 two-drug-resistant strains; 26 three-drug-resistant strains; 14 four-drug-resistant strains; 14 five-drug-resistant strains; 4 six-drug-resistant strains; 3 seven-drug-resistant strains; and especially, one strain isolated from egg product resistant to 9 antibiotic agents. Based on the data, 87 (87/93, 93.55%) isolates were resistant to more than one antibiotic, and 62 (62/93, 66.67%) were multidrug-resistant (≥3 antimicrobial agents). In all the 93 strains, a large proportion of the isolates were resistant to penicillin (86/93, 92.47%), followed by clindamycin (73/93, 78.49%), erythromycin (54/93, 58.06%), tetracycline (24/93, 25.81%), kanamycin (24/93, 25.81%), chloramphenicol (19/93, 20.43%), oxacillin (14/93, 15.05%), ciprofloxacin (10/93, 10.75%), cefoxitin (9/93, 9.68%), nitrofurantoin (5/93, 5.38%), gentamicin (2/93, 2.15%) and mupirocin (2/93, 2.15%) (). In addition, the frequency of intermediate resistance to clindamycin, erythromycin, kanamycin, chloramphenicol, ciprofloxacin, and nitrofurantoin was 44.31%, 23.66%, 15.05%, 11.83%, 9.68% and 3.23% respectively. All 93 strains were sensitive to imipenem and vancomycin ().
Table 4. The profile of antimicrobial susceptibility pattern, antibiotic resistance genes and se/sel genes for 93 S. aureus isolated from food.
Tabla 4. Perfil del patrón de susceptibilidad antimicrobiana, genes de resistencia a los antibióticos y genes se/sel de 93 cepas de S. aureus aisladas de alimentos.
The profile of antibiotic resistance genotypes
In all the 93 food isolates, 50 strains (50/93, 53.76%) carried 10 kinds of antibiotic resistance genes (). Among the 14 resistant genes, the presence of norA (23/93, 24.73%), chlA (23/93, 24.73%) and grlA (22/93, 23.66%) genes was found to be relatively high; followed by ermC (13/93, 13.98%), tetA (11/93, 11.83%), aac6ʹ/aph2” (10/93, 10.75%), ermB (9/93, 9.68%), tetM (3/93, 3.22%), msrA (2/93, 2.15%) and blaZ (1/93, 1.08%). The blaTEM, oxa, ermA, mefA, vanA and mecA genes were not found in any of the 93 isolates. Among the strains, 17 strains carried one antibiotic resistance gene; 17 strains carried two antibiotic resistance genes; 4 strains carried three antibiotic resistance genes; 9 strains carried four antibiotic resistance genes and 4 strains carried five antibiotic resistance genes. Therefore, there were 17 isolates harboring three or more than three antibiotic resistance genes.
The distribution of se/sel gene
As shown in , 43 of 93 S. aureus strains carried se/sel genes. Fourteen of them were from raw meat samples; eight from cooked meat; eight from pickles; four from egg products; four from bean products; two from aquatic products; two from grains and one from fresh vegetable. The prevalence of S. aureus food isolates carrying se/sel gene was 46.24% (43/93), and 8 types of 21 se/sel genes were found. All of them, the occurrence of selx gene was the greatest (30/93, 32.26%), then followed by seb gene (15/93, 16.13%), ser gene (14/93, 15.05%), sea gene (10/93, 10.75%), sec gene (8/93, 8.607%) and selp gene (6/93, 6.45%). The prevalence of sell gene (3/93, 3.23%) and set gene (2/93, 2.15%) was relatively low. Other genes (sed, see, seg, seh, sei, ses, selj, selk, selm, seln, selo, selq and selu) were not found in this investigation. The percentage of two or more kinds of se/sel genes was up to 53.49% (23/43). Among them, 8 strains carried two kinds of se/sel genes; 10 strains carried three kinds of se/sel genes; 3 strains carried four kinds of se/sel genes and 2 strains carried five kinds of se/sel genes (). The genotypes of three or more than three se/sel genes were found in 15 isolates with the combination of traditional se genes and newly identified se/sel genes.
Discussion
The extensive use of antibiotics in humans as well as their use for disease prevention and growth promotion in agriculture has led to the emergence of antibiotic-resistant strains (Hamer & Gill, Citation2002; Martínez & Baquerom, Citation2002; Phillips et al., Citation2004). S. aureus has an outstanding ability to acquire resistance to antibiotics, which is considered a major public health concern (Fernández & Hancock, Citation2012; Meemken et al., Citation2013). In this study, we investigated the antimicrobial susceptibility, antibiotic resistance gene and se/sel genotype of 93 S. aureus isolated from different food sources. Our study has provided an insight into the widespread occurrence of multidrug resistance and virulence factors in S. aureus food isolates and highlighted their potential public health hazards.
Being a normal flora in the environment, or skin, and mucous membranes of animals and humans, S. aureus isolates could acquire antibiotics resistance from other organisms, while the misuse of antimicrobial medicines accelerates this phenomenon (Udegbunam et al., Citation2014). Our study showed a diverse resistance profile of these isolates and a great occurrence of the resistance to 14 antimicrobial agents, which probably was a result of the frequent use of these antibiotics. Especially, the high rate of penicillin resistance (92.47% isolates) called for concern about the use of this drug. Similarly, the relatively high detection rates of resistance to clindamycin (78.49%), erythromycin (58.06%), tetracycline (25.81%), kanamycin (25.81%), chloramphenicol (20.43%), oxacillin (15.05%), ciprofloxacin (10.75%) and cefoxitin (9.68%) also might be due to frequent and indiscriminate use of these drugs, resulting in selection pressure and resistance among these isolates. A study performed in Guangxi Zhuang Autonomous Region of China showed the resistance to penicillin was 95.92%, tetracycline 85.71%, chloramphenicol 81.63% and erythromycin 77.55% (Zhuge, Tan, & Li, Citation2015). Another study in Oklahoma of the United States reported the resistance to penicillin was 52.5%, tetracycline 53.5%, kanamycin 25.7% and erythromycin 32.7% (Abdalrahman, Stanley, Wells & Fakhr, Citation2015). Our results were in consistence with these reports. In our study, S. aureus possessed a relative lower antimicrobial resistance frequency to nitrofurantoin (5.38%), gentamicin (2.15%) and mupirocin (2.15%). One of them, mupirocin, is mainly used in the prevention and treatment of MRSA infections, which is the only inhibitor of aminoacyl t-RNA synthetase in clinical use (Rudresh et al., Citation2015). Even though mupirocin was proved to have a good antibacterial effect several years ago, we still found two mupirocin-resistant isolates. Similarly, the finding of mupirocin-resistant strains has also been reported by other researchers (Nakajima, Hitomi, & Kurihara, Citation2011; Ohadian Moghadam, Pourmand, & Aminharati, Citation2014; Pourmand, Yousefi, Salami, & Amini, Citation2014). As a result, the treatment of high-level resistant strains with mupirocin might not be always effective, which would be a warning owing to the possibility of transmission and dissemination of resistant strains in surroundings. Accordingly, an unnecessary use of mupirocin should be avoided as far as possible. It is notable that all detected strains from food sources were sensitive to imipenem and vancomycin.
When tested strains were resistant to three or more than three kinds of antibiotics, they were considered as multiple drug-resistant strains (Bianchi et al., Citation2014; David et al., Citation2013; Falagas, Koletsi, & Bliziotis, Citation2006). A study by Waters et al. (Citation2011) showed that 52% strains of 64 S. aureus strains isolated from meat and poultry were multiple drug resistance. Xing et al. (Citation2016) also observed that 90.5% of strains from goat milk powder processing plants were resistant to at least one antibiotic. Based on our data, 66.67% strains were multidrug-resistant. These studies showed that multidrug-resistant S. aureus related to food environment had become a serious problem. The assessment of the resistance patterns to different drugs of S. aureus isolated from food might give us an insight into the ecological consequences and antibiotic usage in our society at large.
The detection rates of resistance gene in S. aureus isolated from food ranged from 0% to 24.73% as described earlier. But the phenotype and genotype of antibiotic resistance was inconsistent. Similarly, other researchers also demonstrated that the presence of a certain resistance gene was not necessarily an indicator of antibiotic resistance (Xu et al., Citation2014). The inconsistency between phenotype and genotype happened in most S. aureus isolates tested, which could attribute to other undetected resistance genes, or to the influence of other genetic factors, or to the environmental conditions. It was worth mentioning that no mecA-positive MRSA strains were isolated from any food in this investigation. Our present data only reflected that the incidence of mecA-positive MRSA strains in food in this study was relatively low. In the near future, more epidemiological investigations should be conducted to constantly monitor the antibiotic resistance profiles of S. aureus strains isolated from different food sources in a given area.
Moreover, SEs/SEls are the major virulence factors of S. aureus causing diarrhea and vomiting (Argudín, Mendoza, & Rodicio, Citation2010). Most enterotoxigenic S. aureus strains are found to carry traditional enterotoxin sea, seb, sec and sed. Kérouanton et al. (Citation2007) reported the sea gene (53.6%) was found to be the most common enterotoxin in food poisoning, followed by sed (37.5%) and seb (10%). In this study, the enterotoxigenic S. aureus strains accounted for 46.24%, which was corresponding to 59.8% reported by Normanno et al. (Citation2007) and 54.65% reported by Zhuge et al. (Citation2015). The selx gene in this study possessed the highest detection rate of 32.26%. According to other reports, selx is bacterial super antigen causing human and animal immune modulation which is encoded by core genome and its super antigen activity is even higher than Toxic shock syndrome toxin (TSST) (Wilson et al., Citation2011). Data concerning the prevalence of new SE genes in food poisoning-related S. aureus strains were growing, especially in meat and meat products, vegetables and other foods (Aydin, Sudagidan, & Muratoglu, Citation2011). Among the traditional se genes, seb had the highest detection rate in this study, which may be associated with the Sichuan region climate and geographical environment. We found that strains carrying multiple se/sel genes were very popular. The phenomenon of different se/sel genes co-existance within the same S. aureus had also been reported in other studies (Kellermann et al., Citation2007; Sato’o, Omoe, Naito, Ono, & Nakane, Citation2014; Tang et al., Citation2015). Currently, the direct relationship of S. aureus SEs (with demonstrated emetic activity) and SEls (which lack emetic activity or have yet to be tested) with pathogenicity has not always been established, and the reasons for the redundancy of se/sel genes within the same bacterium deserve further attention (Argudín et al., Citation2010). Based on these investigations, food producer should strengthen the management and quality control of food materials and products to reduce food safety risk.
Taken together, we found that the rate of contamination with S. aureus was serious, which was associated with a great variety of foods. The detection rate of se/sel genes was relatively high and the proportion of multidrug resistance strains was on the rise. It should be mentioned that multidrug resistance stains always carried a variety of se/sel genes. The results on the characteristics of S. aureus food isolates would bring some new thoughts for the evolution of S. aureus pathogenicity. Our investigation has provided an insight into the widespread nature of multidrug-resistant S. aureus strains in food and highlights their potential public health hazards. The public authorities should pay more attention to this phenomenon.
Acknowledgements
This work was jointly supported by the National Natural Science Foundation of China (No. 31371781), the Applied Basic Research Programs of Sichuan Province (2014JY0253), the New Century Excellent Talents in University (NCET-11-0847), and the Project Sponsored by the Scientific Research Fosundation for Returned Scholars, Ministry of Education of China.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- [CLSI] Clinical and Laboratory Standards Inst. (2012). Performance standards for antimicrobial susceptibility testing: Twenty-second information supplement M100-S22. Wayne, Pa: CLSI.
- Abdalrahman, L.S., Stanley, A., Wells, H., & Fakhr, M.K. (2015). Isolation, virulence, and antimicrobial resistance of Methicillin-Resistant Staphylococcus aureus (MRSA) and Methicillin Sensitive Staphylococcus aureus (MSSA) Strains from Oklahoma Retail Poultry Meats. International Journal of Environmental Research and Public Health, 12, 6148–6161. doi:10.3390/ijerph120606148
- Aminov, R.I., Garrigues-Jeanjean, N., & Mackie, R.I. (2001). Molecular ecology of tetracycline resistance: Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Applied and Environmental Microbiology, 67, 22–32. doi:10.1128/AEM.67.1.22-32.2001
- Argudín, M.Á., Mendoza, M.C., & Rodicio, M.R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel), 2, 1751–1773. doi:10.3390/toxins2071751
- Aydin, A., Sudagidan, M., & Muratoglu, K. (2011). Prevalence of staphylococcal enterotoxins, toxin genes and genetic-relatedness of foodborne Staphylococcus aureus strains isolated in the Marmara Region of Turkey. International Journal of Food Microbiology, 148, 99–106. doi:10.1016/j.ijfoodmicro.2011.05.007
- Bianchi, D.M., Gallina, S., Bellio, A., Chiesa, F., Civera, T., & Decastelli, L. (2014). Enterotoxin gene profiles of Staphylococcus aureus isolated from milk and dairy products in Italy. Letters in Applied Microbiology, 58, 190–196. doi:10.1111/lam.12182
- Carfora, V., Giacinti, G., Sagrafoli, D., Marri, N., Giangolini, G., Alba, P., … Battisti, A. (2016). Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: An intra-farm study. Journal of Dairy Science, 99, 4251–4258. doi:10.3168/jds.2016-10912
- Chambers, H.F. (2001). The changing epidemiology of Staphylococcus aureus? Emerging Infectious Diseases, 7, 178–182. doi:10.3201/eid0702.010204
- Choi, S.M., Kim, S.H., Kim, H.J., Lee, D.G., Choi, J.H., Yoo, J.H., … Shin, W.S. (2003). Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. Journal of Korean Medical Science, 18, 631–636. doi:10.3346/jkms.2003.18.5.631
- Chudobova, D., Dostalova, S., Blazkova, I., Michalek, P., Ruttkay-Nedecky, B., Sklenar, M., … Adam, V. (2014). Effect of ampicillin, streptomycin, penicillin and tetracycline on metal resistant and non-resistant Staphylococcus aureus. International Journal of Environmental Research and Public Health, 11, 3233–3255. doi:10.3390/ijerph110303233
- David, M.Z., Taylor, A., Lynfield, R., Boxrud, D.J., Short, G., Zychowski, D., … Daum, R.S. (2013). Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. Medical Center. Journal of Clinical Microbiology, 51, 814–819. doi:10.1128/JCM.02429-12
- DeLeo, F.R., & Chambers, H.F. (2009). Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. The Journal of Clinical Investigation, 119, 2464–2474. doi:10.1172/JCI38226
- Falagas, M.E., Koletsi, P.K., & Bliziotis, I.A. (2006). The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. Journal of Medical Microbiology, 55, 1619–1629. doi:10.1099/jmm.0.46747-0
- Farid, A., Naz, I., Ashraf, A., Ali, A., Rehman, A.U., Sarwar, Y., & Haque, A. (2015). Molecular detection of antimicrobial resistance in local isolates of Staphylococcus epidermidis from urinary tract infections in Faisalabad region of Pakistan. EXCLI Journal, 14, 697–705. eCollection 2015. 10.17179/excli2015-294
- Fernández, L., & Hancock, R.E. (2012). Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clinical Microbiology Reviews, 25(4), 661–681. doi:10.1128/CMR.00043-12
- Frana, T.S., Beahm, A.R., Hanson, B.M., Kinyon, J.M., Layman, L.L., Karriker, L.A., … Smith, T.C. (2013). Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One, 8, e53738. doi:10.1371/journal.pone.0053738
- Hamer, D.H., & Gill, C.J. (2002). From the farm to the kitchen table: The negative impact of antimicrobial use in animals on humans. Nutrition Reviews, 60, 261–264. doi:10.1301/002966402320289395
- Jackson, C.R., Davis, J.A., & Barrett, J.B. (2013). Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. Journal of Clinical Microbiology, 51, 1199–1207. doi:10.1128/JCM.03166-12
- Jansen, K.U., Girgenti, D.Q., Scully, I.L., & Anderson, A.S. (2013). Vaccine review: “Staphyloccocus aureus vaccines: Problems and prospects”. Vaccine, 31, 2723–2730. doi:10.1016/j.vaccine.2013.04.002
- Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H., Forey, F., … Vandenesch, F. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infection and Immunity, 70, 631–641. doi:10.1128/IAI.70.2.631-641.2002
- Jernigan, J.A., Clemence, M.A., Stott, G.A., Titus, M.G., Alexander, C.H., Palumbo, C.M., & Farr, B.M. (1995). Control of methicillin-resistant Staphylococcus aureus at a university hospital: One decade later. Infection Control and Hospital Epidemiology, 16, 686–696. doi:10.2307/30141911
- Kejela, T., & Bacha, K. (2013). Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus aureus (MRSA) among primary school children and prisoners in Jimma Town, Southwest Ethiopia. Annals of Clinical Microbiology and Antimicrobials, 12, 11. doi:10.1186/1476-0711-12-11
- Kellermann, M.G., Sobral, L.M., da Silva, S.D., Zecchin, K.G., Graner, E., Lopes, M.A., … Coletta, R.D. (2007). Mutual paracrine effects of oral squamous cell carcinoma cells and normal oral fibroblasts: Induction of fibroblast to myofibroblast transdifferentiation and modulation of tumor cell proliferation. Oral Oncology, 44, 509–517. doi:10.1016/j.oraloncology.2007.07.001
- Kérouanton, A., Hennekinne, J.A., Letertre, C., Petit, L., Chesneau, O., Brisabois, A., & De Buyser, M.L. (2007). Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. International Journal of Food Microbiology, 115, 369–375. doi:10.1016/j.ijfoodmicro.2006.10.050
- Lina, G., Quaglia, A., Reverdy, M.E., Leclercq, R., Vandenesch, F., & Etienne, J. (1999). Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrobial Agents and Chemotherapy, 43, 1062–1066.
- Martínez, J.L., & Baquerom, F. (2002). Interactions among strategies associated with bacterial infection: Pathogenicity, epidemicity, and antibiotic resistance. Clinical Microbiology Reviews, 15, 647–679. doi:10.1128/CMR.15.4.647-679.2002
- Meemken, D., Blaha, T., Hotzel, H., Strommenger, B., Klein, G., Ehricht, R., … Kehrenberg, C. (2013). Genotypic and phenotypic characterization of Staphylococcus aureus isolates from wild boars. Applied and Environmental Microbiology, 79, 1739–1742. doi:10.1128/AEM.03189-12
- Merz, A., Stephan, R., & Johler, S. (2016). Staphylococcus aureus isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Frontiers in Microbiology, 7, 319. eCollection 2016. 10.3389/fmicb.2016.00319
- Monecke, S., Coombs, G., Shore, A.C., Coleman, D.C., Akpaka, P., Borg, M., … Ehricht, R. (2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One, 6, e17936. doi:10.1371/journal.pone.0017936
- Murakami, K., Minamide, W., Wada, K., Nakamura, E., & Teraoka, H. (1991). Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. Journal of Clinical Microbiology, 29, 2240–2244.
- Nakajima, J., Hitomi, S., & Kurihara, Y. (2011). Detection of methicillin-resistant Staphylococcus aureus with high-level resistance to mupirocin. Journal of Infection and Chemotherapy, 17, 868–871. doi:10.1007/s10156-011-0242-1
- Netsvyetayeva, I., Fraczek, M., Piskorska, K., Golas, M., Sikora, M., Mlynarczyk, A., … Iannitti, T. (2014). Staphylococcus aureus nasal carriage in Ukraine: Antibacterial resistance and virulence factor encoding genes. BMC Infectious Diseases, 14, 128. doi:10.1186/1471-2334-14-128
- Normanno, G., La Salandra, G., Dambrosio, A., Quaglia, N.C., Corrente, M., Parisi, A., … Celano, G.V. (2007). Occurrence, characterization and antimicrobial resistance of enterotoxigenic Staphylococcus aureus isolated from meat and dairy products. International Journal of Food Microbiology, 115, 290–296. doi:10.1016/j.ijfoodmicro.2006.10.049
- Ohadian Moghadam, S., Pourmand, M.R., & Aminharati, F. (2014). Biofilm formation and antimicrobial resistance in methicillin-resistant Staphylococcus aureus isolated from burn patients, Iran. Journal of Infection in Developing Countries, 8, 1511–1517. doi:10.3855/jidc.5514
- Phillips, I., Casewell, M., Cox, T., De Groot, B., Friis, C., Jones, R., … Waddell, J. (2004). Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. The Journal of Antimicrobial Chemotherapy, 53, 28–52. doi:10.1093/jac/dkg483
- Pourmand, M.R., Yousefi, M., Salami, S.A., & Amini, M. (2014). evaluation of expression of nora efflux pump in ciprofloxacin resistant Staphylococcus aureus against Hexahydroquinoline Derivative by Real-Time PCR. Acta Medica Iranica, 52, 424–429.
- Puah, S.M., Chua, K.H., & Tan, J.A. (2016). virulence factors and antibiotic susceptibility of staphylococcus aureus isolates in ready-to-eat foods: Detection of S. aureus contamination and a high prevalence of virulence genes. International Journal of Environmental Research and Public Health, 13, 199. doi:10.3390/ijerph13020199
- Roberts, M.C., Sutcliffe, J., Courvalin, P., Jensen, L.B., Rood, J., & Seppala, H. (1999). Nomenclature for macrolide and macrolide lincosamide-streptogramin B resistance determinants. Antimicrobial Agents and Chemotherapy, 43, 2823–2830.
- Rudresh, M.S., Ravi, G.S., Motagi, A., Alex, A.M., Sandhya, P., & Navaneeth, B.V. (2015). Prevalence of mupirocin resistance among staphylococci, its clinical significance and relationship to clinical use. Journal of Laboratory Physicians, 7, 103–107. doi:10.4103/0974-2727.163127
- Sabath, L.D., Wheeler, N., Laverdiere, M., Blazevic, D., & Wilkinson, B.J. (1977). A new type of penicillin resistance of Staphylococcus aureus. Lancet, 1, 443–447. doi:10.1016/S0140-6736(77)91941-9
- Saha, B., Singh, A.K., Ghosh, A., & Bal, M. (2008). Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). Journal of Medical Microbiology, 57, 72–79. doi:10.1099/jmm.0.47144-0
- Sato’o, Y., Omoe, K., Naito, I., Ono, H.K., & Nakane, A. (2014). Molecular epidemiology and identification of a Staphylococcus aureus clone. Journal of Clinical Microbiology, 52, 2637–2640. doi:10.1128/JCM.00661-14
- Schmitz, F.J., Jones, M.E., Hofmann, B., Scheuring, S., Luckefahr, M., Fluit, A., … Kohrer, K. (1998). Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrobial Agents and Chemotherapy, 42, 1249–1252.
- Shanehbandi, D., Baradaran, B., Sadigh-Eteghad, S., & Zarredar, H. (2014). Occurrence of methicillin resistant and enterotoxigenic Staphylococcus aureus in traditional cheeses in the North West of Iran. ISRN Microbiology, 2014, 129580. eCollection 2014. 10.1155/2014/129580
- Simonetti, O., Cirioni, O., Orlando, F., Alongi, C., Lucarini, G., Silvestri, C., … Provinciali, M. (2011). Effectiveness of antimicrobial photodynamic therapy with a single treatment of RLP068/Cl in an experimental model of Staphylococcus aureus wound infection. The British Journal of Dermatology, 164, 987–995. doi:10.1111/j.1365-2133.2011.10232.x
- Srinivasan, V., Sawant, A.A., Gillespie, B.E., Headrick, S.J., Ceasaris, L., & Oliver, S.P. (2006). Prevalence of enterotoxin and toxic shock syndrome toxin genes in Staphylococcus aureus isolated from milk of cows with mastitis. Foodborne Pathogens and Disease, 3, 274–283. doi:10.1089/fpd.2006.3.274
- Sung, J.M., Lloyd, D.H., & Lindsay, J.A. (2008). Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology, 154, 1949–1959. doi:10.1099/mic.0.2007/015289-0
- Tang, J.N., Zhang, R., Chen, J., Zhao, Y.Y., Tang, C., Yue, H., … Shi, H. (2015). Incidence and characterization of Staphylococcus aureus strains isolated from food markets. Annals of Microbiology, 65, 279–286. doi:10.1007/s13213-014-0859-2
- Tang, Q., Song, P., Li, J., Kong, F., Sun, L., & Xu, L. (2016). Control of antibiotic resistance in China must not be delayed: The current state of resistance and policy suggestions for the government, medical facilities, and patients. Bioscience Trends, 10, 1–6. doi:10.5582/bst.2016.01034
- Udegbunam, S.O., Udegbunam, R.I., & Anyanwu, M.U. (2014). Occurrence of staphylococcal ocular infections of food producing animals in nsukka southeast, Nigeria. Veterinary Medicine International, 2014, 528084. doi:10.1155/2014/528084
- Vahaboglu, H., Ozturk, R., Akbal, H., Saribas, S., Tansel, O., & Coşkunkan, F. (1998). Practical approach for detection and identification of OXA-10-derived ceftazidime-hydrolyzing extended-spectrum beta-lactamases. Journal of Clinical Microbiology, 36, 827–829.
- Wang, Q., Tang, J.N., Tang, C., Chen, J., Liu, J., & Cai, Z. (2016). Temporal expression of staphylococcal enterotoxin K (sek) gene in three isolates from food sources. Food Science, 37, 140–146. [In Chinese.]
- Wang, Q., Tang, J.N., Tang, C., Chen, J., Liu, J., & Chen, L.H. (2015). Rapid method with microwave oven heating for bacterial DNA extraction applied to PCR amplification. Journal of Southwest University for Nationalities (Natural Science Edition), 41, 150–155. [In Chinese.]
- Wang, S.J., Chow, L.W., & Wu, M.J. (2002). Multiplex PCR for the simultaneous detection of the SEA, SEB, SEC, SED and SEE genes of enterotoxigenic Staphylococcus aureus. Journal of Food and Drug Analysis, 10, 164–169.
- Waters, A.E., Contente-Cuomo, T., Buchhagen, J., Liu, C.M., Watson, L., Pearce, K., … Price, L.B. (2011). Multidrug-Resistant Staphylococcus aureus in US Meat and Poultry. Clinical Infectious Diseases, 52, 1227–1230. doi:10.1093/cid/cir181
- WHO, (2014). Antimicrobial resistance: global report on surveillance 2014. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en.
- Wilson, G.J., Seo, K.S., Cartwright, R.A., Connelley, T., Chuang-Smith, O.N., Merriman, J.A., … Fitzgerald, J.R. (2011). A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathogens, 7, e1002271. doi:10.1371/journal.ppat.1002271
- Wongboot, W., Chomvarin, C., Engchanil, C., & Chaimanee, P. (2013). Multiplex PCR for detection of superantigenic toxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolated from patients and carriers of a hospital in northeast Thailand. The Southeast Asian Journal of Tropical Medicine and Public Health, 44, 660–671.
- Xing, X.N., Zhang, Y., Wu, Q., Wang, X., Ge, W.P., & Wu, C.M. (2016). Prevalence and characterization of Staphylococcus aureus isolated from goat milk powder processing plants. Food Control, 59, 644–650. doi:10.1016/j.foodcont.2015.06.042
- Xu, J., Shi, C., Song, M., Xu, X., Yang, P., Paoli, G., & Shi, X. (2014). Phenotypic and genotypic antimicrobial resistance traits of foodborne Staphylococcus aureus isolates from Shanghai. Journal of Food Science, 79, M635–642. doi:10.1111/1750-3841.12405
- Yan, X., Song, Y., Yu, X., Tao, X., Yan, J., Luo, F., … Grundmann, H. (2015). Factors associated with Staphylococcus aureus nasal carriage among healthy people in Northern China. Clinical Microbiology and Infection, 21, 157–162. doi:10.1016/j.cmi.2014.08.023
- Zhuge, S.Y., Tan, D.M., & Li, X.G. (2015). Research on Staphylococcus aureus enterotoxin property and drug resistance in quick frozen flour product in Guangxi. Modern Preventive Medicine, 42, 1765–1767. [In Chinese.]