ABSTRACT
Antioxidant and anti-inflammatory effects of seeds and root extracts (PGSE vs. PGRE) prepared from Platycodon grandiflorum (PG) by ethanol extraction were evaluated. PGSE exhibited greater 2,2-diphenyl-l-picrylhydrazyl (DPPH•) and 2,2-azinobis (3-ethyl-benzothiazoline-6-sulfonic acid) (ABTS•+) radical scavenging activities and more effectively inhibited production of nitric oxide (NO) and prostaglandin E2 (PGE2) compare to PGRE. The release of tumor necrosis factor-α (TNF-α) was also decreased by 31% at 100 μg/mL of PGSE in LPS-stimulated RAW 264.7 macrophages. LPS-induced elevated expressions of inducible NO synthase, cyclooxygenase-2, and pro-inflammatory cytokines (interleukin (IL)-1β, IL-4, and IL-6) were significantly downregulated, in a dose-dependent manner, by PGSE treatment. The suppression of nuclear factor κB (NFκB) translocation and stimulation of heme oxygenase-1 induction were related to anti-inflammatory mechanism of PGSE. Based on these findings, PGSE has the potential to be used as a functional agent for the mitigation of inflammation related diseases.
RESUMEN
Se evaluaron los efectos antioxidantes y antiinflamatorios de las semillas y los extractos de raíces (PGSE vs. PGRE) preparados a partir de Platycodon grandiflorum (PG) utilizando etanol para la extracción. En comparación con el PGRE, el PGSE mostró mayor actividad de eliminación de radicales de 2,2-difenil-l-picrilhidrazilo (DPPH•) y 2,2-azinobis (ácido 3-etilo-benzotiazolina-6-sulfónico) (ABTS•+), así como ser un inhibidor más eficaz de la producción de óxido nítrico (NO) y prostaglandina E2 (PGE2). Asimismo, la liberación del factor de necrosis tumoral-α (TNF-α) se redujo en 31% en 100 μg/mL de PGSE en macrófagos RAW 264.7 inducidos por LPS. Como resultado del tratamiento con PGSE, las elevadas expresiones inducidas por el LPS de la sintasa inducible de NO, la ciclooxigenasa-2 y las citoquinas proinflamatorias (interleucina (IL)-1β, IL-4 e IL-6) experimentaron una infra-regulación significativa, de manera dependiente de la dosis. La supresión de la translocación del factor nuclear B κB (NFκB) y la estimulación de la inducción de la hemo-oxigenasa-1 estaban relacionadas con el mecanismo antiinflamatorio del PGSE. A partir de estos hallazgos, se concluye que la PGSE tiene potencial para ser usado como agente funcional en la mitigación de enfermedades a las que se asocian inflamación.
1. Introduction
Inflammation is an immune response initiated to maintain intercellular homeostasis. However, prolonged chronic inflammation has been linked to the development of various autoimmune diseases including atherosclerosis and type I diabetes (Evans et al., Citation2006). In addition, a low level of pro-inflammatory state is closely related to the onset of cardiovascular diseases (Ritchie & Connell, Citation2007). Thus, the screening of safe and effective sources of potential anti-inflammatory agents and understanding of their modes of action, will be essential for their development.
Platycodon grandiflorum (PG) is a perennial herb that grows in Korea, China, and Japan. The root of PG is used for food and as a traditional alternative medicine in Asian countries to alleviate inflammation, allergies, and oxidative stress (Choi et al., Citation2014; Nyakudya et al., Citation2014). Various bioactivities of PG extract including anti-inflammation, liver protection, anti-obesity, and anti-diabetic activity have been reported, and platycodin D, a triterpenoid saponin, has been considered a major active compound in PG (Qin et al., Citation2014; K. S. Kim et al., Citation2000; Zhao et al., Citation2008).
To date, most studies have highlighted bioactivities of PG root (PGR), with only a few scientific reports regarding PG seed extract (PGSE). In a previous study, we demonstrated that total phenolic and total flavonoid contents of PGSE are significantly higher than those of the root extract (PGRE) (Yoon et al., Citation2018). PGSE also yielded a greater glucose uptake promotion effect compared to PGSE in insulin resistance-induced C2C12 muscle cells (Yoon et al., Citation2018). As part of an ongoing study exploring various bioactivities of PGSE, its anti-inflammatory activities and their underlying mechanisms were investigated using RAW 264.7 macrophages.
2. Materials and methods
2.1. Materials
RAW 264.7 macrophage cells were obtained from ATCC (Manassas, VA, USA). Dulbecco’s Modified Eagle’s Medium (DMEM), penicillin-streptomycin and fetal bovine serum from WelGENE Inc. (Daegu, Korea); a prostaglandin E2 (PGE2) assay kit from Cayman Chemical Company (Ann Arbor, MI, USA) and a TNF-α enzyme-linked immunosorbent assay (ELISA) kit was obtained from eBioscience (San Diego, CA, USA); a high capacity RNA-to-cDNA kit, Taqman® Universal master mix, and a Taqman® gene expression assay kit from Applied Biosystems (Foster City, CA, USA); the LPS and all the other reagents and solvents from Sigma-Aldrich Inc. (St. Louis, MO, USA); and the Nuclear Extraction Kit from Active Motif Inc (Carlsbad, CA, USA). All antibodies (HO-1, Nrf-2, NF-κB, β-actin and TBP) were obtained from Cell Signaling Technology (Danvers, MA, USA).
2.2. Preparation of PGSE and PGRE
PG seeds and PG root were harvested in 2018 from the Gyeongnam province in South Korea, were purchased from Aram-Jongmyo (Seoul, Korea) and Hans herb (Yeongcheon, Korea), respectively. PG root was cut into pieces and ground to obtain homogenous sample. PG seeds (200 g) and PG root (200 g) were extracted with ethanol (100%, 2 L) for 6 h using a solvent reflux extraction unit (Daehan Scientific, Seoul, Korea). This extraction procedure was repeated 3 times, and then the extracts were combined. After the removal of the solvent using a rotary evaporator (Eyela, Tokyo, Japan) at 40°C, each extract was lyophilized and stored at −40°C until used for the cell cultures.
2.3. DPPH • and ABTS •+ radical scavenging activity
The 2,2-diphenyl-l-picrylhydrazyl (DPPH•) and 2,2-azinobis (3-ethyl-benzothiazoline-6-sulfonic acid) (ABTS•+) radical scavenging activities were determined using the method of Thaipong et al. (Citation2006) and were expressed as IC50 values.
2.4. Cell culture
RAW 264.7 mouse macrophage cells were maintained and subcultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin as previously described (Lee & Imm, Citation2017). Briefly, RAW 264.7 cells (5X105 cells/well) were seeded and samples (PGSE and PGRE: 50, 100, and 200 μg/mL in 1% DMSO) and LPS (0.5 μg/mL) were treated for 24 h. The supernatant of the cell medium was collected to measure the levels of cytokines, nitric oxide (NO), tumor necrosis factor-α (TNF-α) and PGE2.
2.5. Cell viability (MTT assay)
The cytotoxicity of cells at different sample concentrations was determined using MTT [3- (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. After 3 h incubation with MTT solution the culture medium was removed and the insoluble formazan crystals were dissolved in dimethyl sulfoxide (DMSO). Absorbance at 540 nm was measured using a plate reader (Biotek Instrument Inc., Winooski, VT, USA).
2.6. Nitric oxide (NO)
The levels of nitrite in the culture media were determined by Griess reaction (Marcocci et al., Citation1994). The culture supernatant (100 μL) was mixed with equal volumes of freshly prepared Griess reagent and absorbance at 546 nm was measured using a plate reader. The concentration of nitrite was calculated from the standard curve constructed with sodium nitrite.
2.7. TNF-α and PGE2
The changes in the production of TNF-α and PGE2 were determined in cell culture supernatant after sample treatment for 24 h. The assays were performed using a TNF-α enzyme linked immunosorbent assay (ELISA) kit (eBioscience) and prostaglandin E2 enzyme immunoassay kit (Cayman Chemical Company) according to the manufacturers’ protocols.
2.8. RNA extraction and quantitative real time PCR
RAW 264.7 cells were cultured on a 6-well plate at a concentration of 5 × 105 cells/well for 24 h in the presence of samples and LPS (0.5 μg/mL). Total RNA was extracted using QIAzol® Lysis Reagent (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions, and single-strand cDNA was synthesized using a cDNA kit (Applied Biosystems). The quantitative real-time PCR was carried out as previously described (Lee & Imm, Citation2017). The following primers were used in the analysis: β-actin (Mm00607939_s1), iNOS (Mm00440502_m1), COX2 (Mm00478374_m1), IL-1β (Mm00434228_m1) and IL-6 (Mm00446190_m1). Taqman probes (dual-labeled with 6-carboxyfluorescein as the 5ʹ-reporter and 3ʹ TAMRA quencher) were used for assays.
2.9. Western blotting analysis
Nuclear and cytoplasmic extract were prepared using a Nuclear Extract Kit (Active Motif) containing protease and phosphatase inhibitors (Next Advance, Troy, NY, USA). Equal amounts of protein were separated by SDS-PAGE (10%) and then transferred onto polyvinylidene difluoride membranes. After blocking the membranes with 5% BSA, they were incubated with a specific primary antibodies (1:1000 dilution) overnight at 4°C. They were washed with TBST buffer and then was incubated with a horseradish peroxidase conjugated secondary antibody (1:3000 dilution) at room temperature for 1 h. The protein bands were visualized using an enhanced chemiluminescent detection system and band intensities were quantified using Image Lab software (BIO-RAD, Hercules, CA, USA).
2.10. Statistical analysis
All experiments were carried out in triplicates and the results were expressed as means ± standard deviations. One way analysis of variance (ANOVA) was used to determine the differences between multiple groups. When a significant difference (P < 0.05) was found in ANOVA, Duncan’s multiple comparisons test was conducted. A paired t-test was used to evaluate the differences in radical scavenging activities (DPPH• and ABTS•+) between PGSE and PGRE. All data were analyzed using SPSS 18.0 (Chicago, IL, USA).
3. Results and discussion
3.1. Antioxidant activity of PGSE and PGRE
As shown in , PGSE showed a significantly higher scavenging activity (lower IC50 values) for both DPPH• and ABTS•+ when compared to PGRE (p < 0.05). DPPH• and ABTS•+ radical scavenging activities have been widely used as an indices for antioxidant activities of foods. ABTS•+ radical scavenging activity can be applied regardless of antioxidant polarity; however, DPPH• assay is more acceptable for the evaluation of hydrophobic antioxidants since it uses DPPH• radicals that have been dissolved in organic media (D.-O. Kim et al., Citation2002). It is well known that phenolic compounds possess strong antioxidant activity, notably radical scavenging activity. The high radical scavenging activity of samples showed strong correlation with phenolic content in Nigella staiva seeds (Khattak et al., Citation2008).
Table 1. DPPH and ABTS radical scavenging activities of PGSE and PGRE.
Tabla 1. Actividades de eliminación de radicales de DPPH y ABTS de PGSE y PGRE.
Yoon et al. (Citation2018) reported that PGSE had greater polyphenols (243 vs. 26 mg gallic acid equivalent/g extract) and flavonoids (124 vs. 14 mg catechin equivalent/g extract) levels compared to PGRE. However, there was no significant difference in the total saponin content between PGSE and PGRE. Wang et al. (Citation2017) analyzed the phytochemical constituents from different parts of PG plants grown in northeast China. They found that the seed contained more flavonoids content but lower saponins than those found in the root. Some discrepancies between the saponin contents of PGSE and PGRE may be ascribed to conditions during extraction. The optimal condition for saponin extraction is a temperature of 40°C using 50–70% ethanol rather than 100% ethanol (Chen et al., Citation2009; Ngo et al., Citation2017).
Based on our previous results, the greater radical scavenging activities of PGSE are mainly due to greater total phenolic and flavonoid contents. Jeong et al. (Citation2010) reported that butanol extract from the aerial parts of PG displayed stronger antioxidant activity than chloroform and water extracts with luteolin-7-O-glucoside and apigenin-7-O-glucoside being predominant compounds present in the butanol fraction.
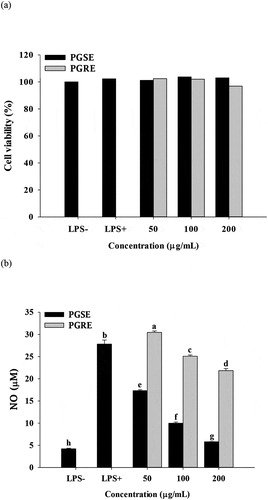
3.2. Effect of PGSE and PGRE on NO production
No cytotoxicity was found in up to 200 μg/mL of PGSE or PGRE (), and the anti-inflammatory activities of PGSE were evaluated within the non-cytotoxic concentration range. NO increased up to 27 μM in response to LPS-stimulation. It was significantly decreased, in a dose-dependent manner, by PGSE or PGRE treatment (). Overall, PGSE displayed a much stronger inhibitory effect compared to PGRE on NO production in LPS-stimulated RAW 264.7 cells. In our previous study, luteolin was identified as an active compound of PGSE which improves palmitate-induced insulin resistance in C2C12 muscle cells. Approximately 38% of total phenolics in PGSE are luteolin (Yoon et al., Citation2018). Xagorari et al. (Citation2001) reported that luteolin actively suppressed NO and other pro-inflammatory cytokines in LPS-induced RAW 264.7 cells. They pointed out that luteolin was more potent as an anti-inflammatory agent compared to luteolin-7-O-glucoside, since the increased aqueous solubility of luteolin glycoside possibly limited its affinity for cellular membranes. In addition, Ahn et al. (Citation2005) reported that PG saponins including 2”-O-acetyl polygalacin, platycodin A, and platycodin D decreased NO production in LPS-stimulated RAW 264.7 cell.
Figure 1. Effects of PGSE and PGRE on cell viability (a) and NO production (b) of LPS-stimulated RAW 264.7 cells. PGSE: Platycodon grandiflorum seed extract, PGRE: Platycodon grandiflorum root extract, NO: nitric oxide. Different letters indicate significant differences at p < 0.05.
Figura 1. Efectos de PGSE y PGRE en la viabilidad celular (a) y producción de NO (b) de células RAW 264.7 estimuladas por LPS.PGSE: Extracto de semilla de Platycodon grandiflorum, PGRE: Extracto de raíz de Platycodon grandiflorum, NO: óxido nítrico. Las distintas letras indican diferencias significativas en p < 0.05.
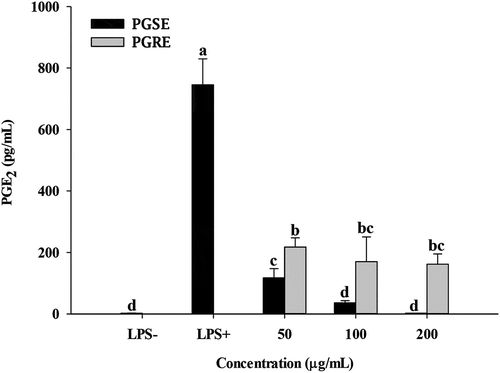
3.3. Effects of PGSE and PGRE on the production of PGE2
As shown in , the production of PGE2 was significantly decreased in a dose-dependent manner. When 200 μg/mL PGSE was treated, the level of PGE2 production was similar to the basal level of control. A significant difference was found in the level of PGE2 between PGSE and PGRE at all the tested concentrations (). Prostaglandins are oxygenated 20 carbon fatty acid groups that are synthesized by the conversion of arachidonic acid through inflammatory stimuli. COX is the first enzyme to synthesize prostaglandins and synthesizes PGE2 through complex binding reactions. The amount of PGE2 increases in inflammatory conditions and causes an acute inflammatory response (Frederick et al., Citation1980). K.-J. Jang et al. (Citation2013) reported that treatment with PG saponins (400 and 500 μg/mL) significantly reduced the release of PGE2 in LPS-stimulated microgilial cells. Results showed that PGSE had greater anti-inflammatory activity than PGRE for the production of pro-inflammatory mediators such as NO and PGE2. Thus, further study was conducted using PGSE.
Figure 2. Effects of PGSE on the production of PGE2 in LPS-stimulated RAW 264.7 macrophages. PGSE: Platycodon grandiflorum seed extract, PGE2: prostaglandin E2. Different letters indicate significant differences at p < 0.05.
Figura 2. Efectos de PGSE en la producción de PGE2 en macrófagos RAW 264.7 estimulados por LPS.PGSE: extracto de semilla de Platycodon grandiflorum, PGE2: prostaglandina E2. Las distintas letras indican diferencias significativas en p < 0.05.
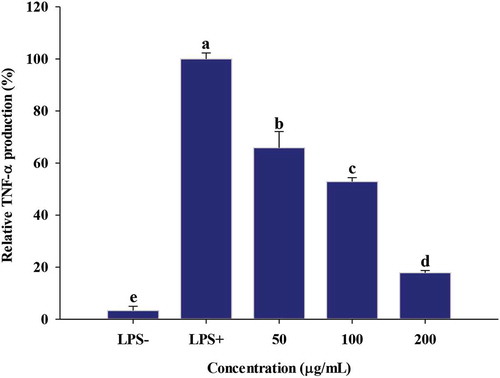
3.4. Effect of PGSE on TNF-α production
Cytokine is a protein that binds to specific cell receptors and modulates the activity and growth of immune cells (E. Y. Kim & Moudgil, Citation2008). TNF-α, a pro-inflammatory cytokine, is cytokine secreted in the early stages of inflammatory response by LPS-stimulated macrophages; it triggers IL-1β and IL-6 expression (Grossman et al., Citation1989). Thus, the suppression of TNF-α plays a key role in the initiation and maintenance of inflammation, especially in chronic inflammatory diseases (Y. J. Kim & Son, Citation2012). As shown in , TNF-α production was increased in response to LPS stimulation, but its levels were decreased by 66%, 53% and 18%, by addition of 50, 100, 200 μg/mL of PGSE, respectively (). The significant reduction of TNF-α by PGSE suggests that the LPS-induced inflammatory response has been effectively downregulated since TNF-α is a key stimulator of the pro-inflammatory signaling cascade and activates inflammatory genes (Sethi et al., Citation2008).
Figure 3. Effect of PGSE on TNF-α production in LPS-stimulated RAW 264.7 macrophages. PGSE: Platycodon grandiflorum seed extract. TNF-α: tumor necrosis factor-α. Different letters indicate significant differences at p < 0.05.
Figura 3. Efecto de PGSE en la producción de TNF-α en los macrófagos RAW 264.7 estimulados por LPS.PGSE: Extracto de semilla de Platycodon grandiflorum. TNF-α: factor de necrosis tumoral-α. Las distintas letras indican diferencias significativas en p < 0.05.
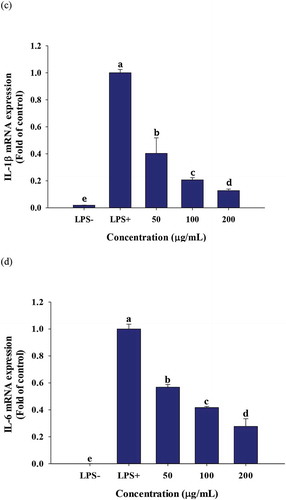
3.5. The effect of PGSE on pro-inflammatory gene expressions
Increased TNF-α production activates various transcription factors involved in the inflammatory response. The effects of PGSE treatment on pro-inflammatory gene expressions were analyzed using quantitative RT-PCR. All pro-inflammatory gene expressions were significantly decreased when treated with 100 and 200 μg/mL of PGSE ().
Figure 4. Effects of PGSE on the expression of iNOS (a), COX2 (b), IL-1β (c), and IL-6 (d) in LPS-stimulated RAW 264.7 macrophages. PGSE: Platycodon grandiflorum seed extract, iNOS: inducible NO synthase, COX2: cyclooxygenase 2, IL-1β: interleukin-1β, IL-6: interleukin-6. Different letters indicate significant differences at p < 0.05.
Figura 4. Efectos del PGSE en la expresión de iNOS (a), COX2 (b), IL-1β (c), y IL-6 (d) en macrófagos RAW 264.7 estimulados por LPS.PGSE: Extracto de semilla de Platycodon grandiflorum, iNOS: sintasa inducible NO, COX2: ciclooxigenasa 2, IL-1β: interleucina-1β, IL-6: interleucina-6. Las distintas letras indican diferencias significativas en p < 0.05.
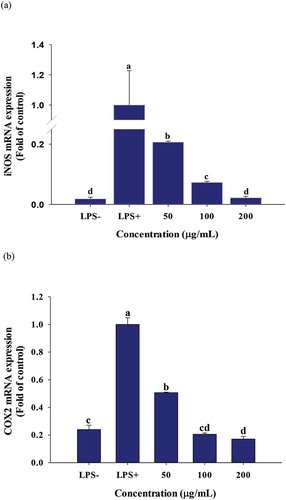
The two inducible enzymes, inducible NO synthase (iNOS) and cyclooxygenase 2 (COX2), are expressed by LPS stimulation; the synthesis of PGE2 from arachidonic acid is mediated by COX2 (Coleman, Citation2001). A cross talk between iNOS and COX2 has been suggested and iNOS inhibitors effectively suppressed inflammation by dual inhibition of NO and PGE2 (Salvemini et al., Citation1995). In this context, the dose-dependent inhibitions of NO and PGE2 observed in and , were due to transcriptional suppression of iNOS and COX2. M. H. Jang et al. (Citation2006) reported that the mRNA expressions of COX2 and iNOS were significantly decreased in LPS-treated microglial BV2 cells in response to 1 mg/mL of water-soluble PG extract. Like TNF-α, IL-1β and IL-6 are primary cytokines that initiate acute inflammation in macrophages. The increased release of IL-1β and IL-6 promotes fever and inflammatory reactions, which cause tissue damage (Duque & Descoteaux, Citation2014).
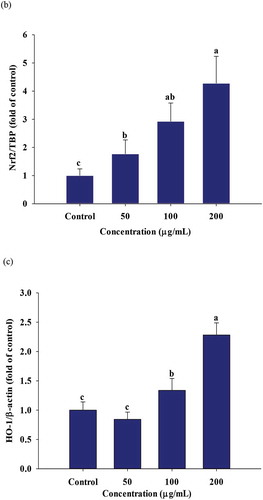
3.6. Effects on PGSE on nuclear factor κB (NFκB) translocation and induction of nuclear factor erythroid 2-related factor 2 (Nrf2) and heme oxygenase-1 (HO-1)
Following LPS stimulation in macrophages, NFκB signals initiate the inflammation process (Li & Verma, Citation2002). Thus, western blotting was used to analyze PGSE-mediated nuclear NFκB translocation and induction of the anti-inflammatory enzyme, heme oxygenase-1 (HO-1). As shown in , NFκB levels translocated into the nuclei were significantly decreased at 100 and 200 μg/mL of PGSE in LPS-stimulated RAW 264.7 cells.
Figure 5. Effects of PGSE on the expression of NFκB (a), Nrf2 (b), and HO-1 (c) in LPS-stimulated RAW 264.7 macrophages. PGSE: Platycodon grandiflorum seed extract, NFκB: nuclear factor κB, Nrf2: nuclear factor erythroid 2-related factor 2, HO-1: heme oxygenase-1. Different letters indicate significant differences at p < 0.05.
Figura 5. Efectos del PGSE en la expresión de NFκB (a), Nrf2 (b), y HO-1 (c) en macrófagos RAW 264.7 estimulados por LPS.PGSE: Extracto de semilla de Platycodon grandiflorum, NFκB: factor nuclear κB, Nrf2: factor nuclear eritrocito 2 relacionado con el factor 2, HO-1: hemo-oxigenasa-1. Las distintas letras indican diferencias significativas en p < 0.05.
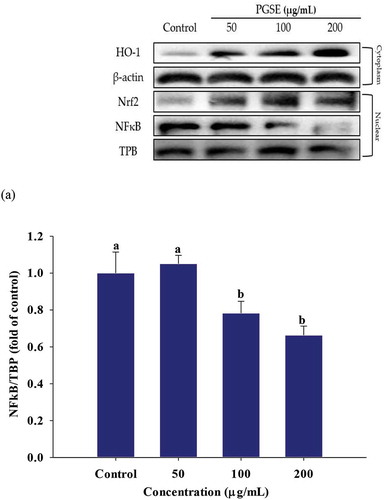
Nrf2 is a transcription factor in cytoplasm and the amount of free Nrf2 increases under oxidative or inflammatory stress. The nuclear Nrf2 translocation leads to its binding to promoters of antioxidant response elements, It also activates the expressions of antioxidant and anti-xenobiotic enzymes including HO-1, quinone reductase, and glutathione S-transferase (Luo et al., Citation2011). The nuclear Nrf2 and HO-1 expressions were significantly increased in a dose-dependent manner, in response to PGSE treatment (,).
HO-1 is expressed under cellular stress, such as oxidative and inflammatory conditions, and catalyzes the conversion of free heme into ferritin, carbon monoxide (CO), and bilirubin. Both heme degradation products and the cell signaling of CO are responsible for the anti-inflammatory effect of HO-1 (Pae & Chung, Citation2009). Kapturczak et al. (Kapturczak et al., Citation2004) demonstrated that HO-1 is an important contributor in the modulation of inflammation by showing the increased pro-inflammatory state was highly associated with HO-1 deficiency in the HO-1-deficient mice model. Sung and Lee (Citation2015) reported that luteolin resulted in the suppression of NFκB translocation and stimulated HO-1 expression in LPS-stimulated RAW 264.7 macrophages. The strong reduction of NFκB-mediated inflammation by PGSE suggests that PGSE it has great potential for alleviating diverse inflammatory diseases.
4. Conclusion
GSE showed greater antioxidant activity compared to PGRE and more effectively inhibited the production of pro-inflammatory biomarkers, including NO and PGE2. LPS-induced elevated expressions of iNOS and COX-2 and pro-inflammatory cytokines in RAW 264.7 cells were significantly downregulated by PGSE treatment. Since PGSE is a mixture of phytochemicals including flavonoids and saponins, multiple anti-inflammatory mechanisms are probably involved. The suppression of nuclear NFκB translocation and the stimulation of HO-1 induction were related to the anti-inflammatory mechanisms of PGSE.
Disclosure statement
The authors declare that they have no potential conflict of interest.
Additional information
Funding
References
- Ahn, K. S., Noh, E. J., Zhao, H. L., Jung, S. H., Kang, S. S., & Kim, Y. S. (2005). Inhibition of inducible nitric oxide synthase and cyclooxygenase II by Platycodon grandiflorum saponins via suppression of nuclear factor-κB activation in RAW 264.7 cells. Life Science, 76(20), 2315–2328. https://doi.org/10.1016/j.lfs.2004.10.042
- Chen, R., Meng, F., Zhang, S., & Liu, Z. (2009). Effects of ultrahigh pressure extraction conditions on yields and antioxidant activity of ginsenoside from ginseng. Separation and Purification Technology, 66(2), 340–346. https://doi.org/10.1016/j.seppur.2008.12.026
- Choi, J. H., Jin, S. W., Han, E. H., Park, B. H., Kim, H. G., Khanal, T., Hwang, Y. P., Do, M. T., Lee, H.-S., Chung, Y. C., Kim, H. S., Jeong, T. C., & Jeong, H. G. (2014). Platycodon grandiflorum root-derived saponins attenuate atopic dermatitis-like skin lesions via suppression of NF-κB and STAT1 and activation of Nrf2/ARE-mediated heme oxygenase-1. Phytomedicine, 21(8–9), 1053–1061. https://doi.org/10.1016/j.phymed.2014.04.011
- Coleman, J. W. (2001). Nitric oxide in immunity and inflammation. International Immunopharmacology, 1(8), 1397–1406. https://doi.org/10.1016/S1567-5769(01)00086-8
- Duque, G. A., & Descoteaux, A. (2014). Macrophage cytokines: Involvement in immunity and infectious diseases. Frontiers in Immunology, 5, 491. https://ind01.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdoi.org%2F10.3389%2Ffimmu.2014.00491&data=02%7C01%7Caravind.devanathan%40integra.co.in%7C9f53a655728140a7f5eb08d8053ce7ce%7C70e2bc386b4b43a19821a49c0a744f3d%7C0%7C0%7C637265106443434709&sdata=kMeJ8nmFTirxrwnySopGczgd1Pv%2FoHCroX%2BlVjSie5M%3D&reserved=0
- Evans, D. A., Hirsch, J. B., & Dushenkov, S. (2006). Phenolics, inflammation and nutrigenomics. Journal of the Science of Food and Agriculture, 86(15), 2503–2509. https://doi.org/10.1002/jsfa.2702
- Frederick, A., Kuehl, Jr., & Egan, R. W. (1980). Prostaglandins, arachidonic acid, and inflammation. Science, 210(4473), 978–984. https://doi.org/10.1126/science.6254151
- Grossman, R. M., Krueger, J., Yourish, D., Granelli-Piperno, A., Murphy, D. P., May, L. T., Kupper, T. S., Sehgal, P. B., & Gottlieb, A. B. (1989). Interleukin-6 is expressed in high levels in psoriatic skin and stimulates proliferation of cultured human keratinocytes. Proceedings of the National Academy of Sciences, 86(16), 6367–6371. https://doi.org/10.1073/pnas.86.16.6367
- Jang, K.-J., Kim, H. K., Han, M. H., Oh, Y. N., Yoon, H.-M., Chung, Y. H., Kim, G. Y., Hwang, H. J., Kim, B. W., & Choi, Y. H. (2013). Anti-inflammatory effects of saponins derived from the roots of Platycodon grandiflorus in lipopolysaccharide-stimulated BV2 microglial cells. International Journal of Molecular Medicine, 31(6), 1357–1366. https://doi.org/10.3892/ijmm.2013.1330
- Jang, M. H., Kim, C. J., Kim, E. H., Kim, M. G., Leem, K. H., & Kim, J. (2006). Effects of Platycodon grandiflorum on lipopolysaccharide-stimulated production of prostaglandin E2, nitric oxide, and interleukin-8 in mouse microglial BV2 cells. Journal of Medicinal Food, 9(2), 169–174. https://doi.org/10.1089/jmf.2006.9.169
- Jeong, C. H., Choi, G. N., Kim, J. H., Kwak, J. H., Kim, D. O., Kim, Y. J., & Heo, H. J. (2010). Antioxidant activities from the aerial parts of Platycodon grandiflorum. Food Chemistry, 118(2), 278–282. https://doi.org/10.1016/j.foodchem.2009.04.134
- Kapturczak, M. H., Wasserfall, C., Brusko, T., Campbell-Thompson, M., Ellis, T. M., Atkinson, M. A., & Agarwal, A. (2004). Heme oxygenase-1 modulates early inflammatory responses: Evidence from the heme oxygenase-1-deficient mouse. The American Journal of Pathology, 165(3), 1045–1053. https://doi.org/10.1016/S0002-9440(10)63365-2
- Khattak, F. K., Simpson, T. J., & Ihasnullah. (2008). Effect of gamma irradiation on the extraction yield, total phenolic content and free radical-scavenging activity of Nigella staiva seed. Food Chemistry, 110(4), 967–972. https://doi.org/10.1016/j.foodchem.2008.03.003
- Kim, D.-O., Lee, K. W., Lee, H. J., & Lee, C. Y. (2002). Vitamin C equivalent antioxidant capacity (VCEAC) of phenolic phytochemicals. Journal of Agricultural and Food Chemistry, 50(13), 3713–3717. https://doi.org/10.1021/jf020071c
- Kim, E. Y., & Moudgil, K. D. (2008). Regulation of autoimmune inflammation by pro-inflammatory cytokines. Immunology Letters, 120(1–2), 1–5. https://doi.org/10.1016/j.imlet.2008.07.008
- Kim, K. S., Seo, E. K., Lee, Y. C., Lee, T. K., Cho, Y. W., Ezaki, O., & Kim, C. H. (2000). Effect of dietary Platycodon grandiflorum on the improvement of insulin resistance in obese Zucker rats. Journal of Nutritional Biochemistry, 11(9), 420–424. https://doi.org/10.1016/S0955-2863(00)00098-X
- Kim, Y. J., & Son, D. Y. (2012). Antioxidant and inhibitory effects of Korean Panax ginseng extract on pro-inflammatory mediators in LPS-stimulated RAW264. 7 macrophages. Journal of the Korean Society of Food Science and Nutrition, 41(10), 1371–1377. https://doi.org/10.3746/jkfn.2012.41.10.1371
- Lee, D., & Imm, J. Y. (2017). AMP kinase activation and inhibition of nuclear factor‐Kappa B (NF‐κB) translocation contribute to the anti‐inflammatory effect of tricin. Journal of Food Biochemistry, 41(2), e12293. https://doi.org/10.1111/jfbc.12293
- Li, Q., & Verma, I. M. (2002). NF-kappa B regulation in the immune system. Nature Reviews. Immunology, 2(10), 725–734. https://doi.org/10.1038/nri910
- Luo, C., Urgard, E., Vooder, T., & Metspalu, A. (2011). The role of COX-2 and Nrf2/ARE in anti-inflammation and antioxidative stress: Aging and anti-aging. Medical Hypotheses, 77(2), 174–178. https://doi.org/10.1016/j.mehy.2011.04.002
- Marcocci, L., Packer, L., Droy-Lefaiz, M. T., Sekaki, A., & Gardes- Albert, M. (1994). Antioxidant action of Ginkgo biloba extracts EGb 761. Methods in Enzymology, 234, 75–84. https://ind01.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdoi.org%2F10.1016%2F0076-6879&data=02%7C01%7Caravind.devanathan%40integra.co.in%7C9f53a655728140a7f5eb08d8053ce7ce%7C70e2bc386b4b43a19821a49c0a744f3d%7C0%7C0%7C637265106443444706&sdata=98EuU8LCnZ9d%2FHyYNODBIH7XiUYXuRVYeEsSQYYqrHw%3D&reserved=0(94)34117-6
- Ngo, T. V., Scarlett, C. J., Bowyer, M. C., Ngo, P. D., & Vuong, Q. V. (2017). Impact of different extraction solvents on bioactive compounds and antioxidant capacity from the root of Salacia chinensis L. Journal of Food Quality Article, 462–475. Article ID 9305047. https://ind01.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdoi.org%2F10.1155%2F2017%2F9305047&data=02%7C01%7Caravind.devanathan%40integra.co.in%7C9f53a655728140a7f5eb08d8053ce7ce%7C70e2bc386b4b43a19821a49c0a744f3d%7C0%7C0%7C637265106443444706&sdata=pWfDQspJRjaUp4rQOV05uo86nNNf9rzmkK%2FUBh9BNTc%3D&reserved=0
- Nyakudya, E., Jeong, J. H., Lee, N. K., & Jeong, Y.-S. (2014). Platycosides from the roots of Platycodon grandiflorum and their health benefits. Preventive Nutrition and Food Science, 19(2), 59–68. https://doi.org/10.3746/pnf.2014.19.2.059
- Pae, H. O., & Chung, H. T. (2009). Heme oxygenase-1: Its therapeutic roles in inflammatory diseases. Immune Network, 9(1), 12–19. https://doi.org/10.4110/in.2009.9.1.12
- Qin, H., Du, X., Zhang, Y., & Wang, R. (2014). Platycodin D, a triterpenoid saponin from Platycodon grandiflorum, induces G2/M arrest and apoptosis in human hepatoma HepG2 cells by modulating the PI3K/Akt pathway. Tumor Biology, 35(2), 1267–1274. https://doi.org/10.1007/s13277-013-1169-1
- Ritchie, S. A., & Connell, J. M. C. (2007). The link between abdominal obesity, metabolic syndrome and cardiovascular disease. Nutrition, Metabolism and Cardiovascular Diseases, 17(4), 319–326. https://doi.org/10.1016/j.numecd.2006.07.005
- Salvemini, D., Settle, S., Masferrer, J., Seibert, K., Currie, M., & Needleman, P. (1995). Regulation of prostag-landin production by nitric oxide; an in vivo analysis. British Journal of Pharmacology, 114(6), 1171–1178. https://doi.org/10.1111/j.1476-5381.1995.tb13330.x
- Sethi, G., Sung, B., & Aggarwal, B. B. (2008). TNF: A master switch for inflammation to cancer. Frontiers Bioscience, 13(2), 5094–5107. https://doi.org/10.2741/3066
- Sung, J., & Lee, J. (2015). Anti-inflammatory activity of butein and luteolin through suppression of NF κ B activation and induction of heme oxygenase-1. Journal of Medicinal Food, 18(5), 557–564. https://doi.org/10.1089/jmf.2014.3262
- Thaipong, K., Boonprakob, U., Crosby, K., Cisneros-Zevallos, L., & Byrne, D. H. (2006). Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. Journal of Food Composition and Analysis, 19(6–7), 669–675. https://doi.org/10.1016/j.jfca.2006.01.003
- Wang, C., Zhang, N., Wang, Z., Qi, Z., Zhu, H., Zheng, B., & Liu, J. (2017). Nontargeted metabolomic analysis of four different parts of Platycodon grandiflorum grown in northeast China. Molecules, 22(8), 1280. https://doi.org/10.3390/molecules22081280
- Xagorari, A., Papapetropoulos, A., Mauromatis, A., Economou, M., Fotsis, T., & Roussos, C. (2001). Luteolin inhibits an endotoxin-stimulated phosphorylation cascade and proinflammatory cytokine production in macrophages. Journal of Pharmacology, 296(1), 181–187. 0022-3565/01/2961-181–187$3.00
- Yoon, H. J., Bang, M. H., Kim, H., & Imm, J. Y. (2018). Improvement of palmitate-induced insulin resistance in C2C12 skeletal muscle cells using Platycodon grandiflorum seed extracts. Food Bioscience, 25, 61–67. https://doi.org/10.1016/j.fbio.2018.08.002
- Zhao, H. L., Harding, S. V., Marinangeli, C. P. F., Kim, Y. S., & Jones, P. J. H. (2008). Hypocholesterolemic and anti‐obesity effects of saponins from Platycodon grandiflorum in hamsters fed atherogenic diets. Journal of Food Science, 73(8), 195–200. https://doi.org/10.1111/j.1750-3841.2008.00915.x