ABSTRACT
Streptococcus suis can cause severe infections in pigs and humans. The tonsils of pigs are major niches for S. suis, and different serotypes of S. suis can be found in the same tonsil. Pig tonsil colonization by S. suis is believed to be an important source of infection for humans and pigs. However, how S. suis competes for a stable tonsil niche is unknown. Here, we found that S. suis strain WUSS351, isolated from a healthy pig tonsil, is virulent and multidrug-resistant. The ABC transporter system SstFEG, conferring resistance to bacitracin, was reported to confer a competitive survival advantage in vivo. In addition, strain WUSS351 has several antimicrobial systems, including a novel type VII secretion system (T7SS), lantibiotic bacteriocin, and lactococcin972-like bacteriocin Lcn351. Bacterial competition experiments demonstrated T7SS-mediated cell contact-dependent antagonism of S. suis. Antibacterial activity analysis and 16S rRNA gene sequencing of the culture-independent and culture-dependent pig tonsillar microbiome revealed that Lcn351 mainly targets S. suis, one of the core microbiomes in pig tonsils. Taken together, our results revealed the mechanism of the stable persistence of S. suis in the tonsil niche, which might have important implications for S. suis epidemiology, potentially influencing strain prevalence and disease progression.
Introduction
Streptococcus suis is able to cause meningitis, septicemia, arthritis, and sudden death in pigs [Citation1]. S. suis is also pathogenic toward humans, acting as a zoonotic agent and infecting via contact with contaminated by-products or infected pigs [Citation2,Citation3]. In addition, S. suis promotes antimicrobial resistance by spreading resistance genes to other streptococci [Citation4]. S. suis is a heterogeneous bacterial species. To date, 29 serotypes have been identified according to the antigenicity of their capsular polysaccharides (CPS). In addition, 28 novel cps loci (NCL) genotypes have been found recently [Citation5–9]. Compared with the 29 serotypes, the information on NCL strains is very limited. The pig tonsil is colonized by many microbes, and it is a reservoir niche for some important bacterial pathogens, such as S. suis [Citation10,Citation11]. In fact, different serotypes of S. suis can be found from the same tonsil [Citation12]. S. suis strains isolated from healthy pig tonsils can be pathogenic [Citation3]. Liang et al. reported that four representative strains of S. suis serotype 7 isolated from healthy pig tonsils were identified as virulent strains; two of them belonged to ST373, the same sequence type with serotype 7 human isolate GX69, suggesting we should be aware of the public health threat posed by S. suis serotype 7 ST373 strains [Citation13]. Wang et al. reported the pathogenicity of three representative strains of S. suis serotype 31 isolated from healthy pig tonsils [Citation14]. Thus, infections in susceptible pigs [Citation15] and humans [Citation16,Citation17] might derive from S. suis in the tonsils of healthy pigs.
In Gram-positive bacteria, the type VII secretion system (T7SS) and bacteriocins are important antimicrobial systems that mediate bacterial competition and infection. T7SS is specific secretion machinery for achieving contact-dependent toxins transport to target prey bacteria [Citation18] and plays an essential role in virulence [Citation19,Citation20] and colonization [Citation21,Citation22] of pathogenic bacteria. Bacteriocins are antimicrobial effectors which play a critical role in bacterial competition to fit its environmental niche [Citation23,Citation24]. However, we do not know how S. suis competes for a stable tonsil niche. The present study reveals the complete genome sequence of S. suis NCL4 strain WUSS351, a virulent and multidrug-resistant strain isolated from a healthy pig tonsil. In addition, this strain has several antimicrobial systems, including T7SS, lantibiotic bacteriocin, and lactococcin972-like bacteriocin Lcn351, which together promote its niche competition in pig tonsils. These findings contribute to understanding how S. suis stably persists in the tonsil niche and help to develop novel ways to control the disease caused by this bacterium.
Materials and methods
Bacterial strains and culture conditions
S. suis strain WUSS351 was isolated from a healthy pig tonsil in Jiangsu province, China. Table S1 provides detailed information on the strains and plasmids. S. suis strains were cultured in Todd-Hewitt broth (THB, Becton Dickinson, Franklin Lakes, NJ, USA) or on 6% (vol/vol) sheep blood-containing agar medium, at 37°C with 5% CO2. For S. suis mutant selection, the medium contained 100 μg/mL spectinomycin (Spc, Sigma-Aldrich, St. Louis, MO, USA), or 5 μg/mL chloramphenicol (Chl, Sigma-Aldrich, St. Louis, MO, USA). Luria-Bertani (LB, Becton Dickinson) medium was used to culture Escherichia coli strains at 37°C. To select bacteria harboring recombinant plasmids, 100 μg/mL ampicillin (Amp, Sigma-Aldrich) or 50 μg/mL Spc were added when necessary.
Genome sequencing and bioinformatic analysis
A Bacterial DNA Kit (OMEGA Biotech, Norcross, GA, USA) was used to extract the genomic DNA from S. suis strain WUSS351, following the manufacturer’s guidelines. Novogene Bioinformatics Technology Co., Ltd (Beijing, China) used the Illumina NovaSeq PE150 system (Illumina, San Diego, CA, USA) to sequence the genomic DNA. SOAP denovo, version 2.04 [Citation25] was used to carry out the de novo genome assembly. We deposited the genome sequence and annotation for strain WUSS351 at NCBI (Accession No. NZ_CP039462.1). Possible genomic islands (GIs) in the genome were predicted using Island Viewer 4 [Citation26]. BLAST algorithms at the NCBI were used to perform homology analysis. The genetic distance among S. suis strains was calculated using the OrthoANIu algorithm [Citation27]. Based on the distance data, a phylogenetic tree by the Neighbor-Joining method was generated with MEGA X software [Citation28]. Multiple alignments of genomes were carried out using the Mauve software [Citation29], and the Circos software was used to display the comparison results [Citation30].
Transmission electron microscopy (TEM)
The capsule of S. suis was observed using TEM using a previously published method [Citation6]. A JEM-1010 TEM (JEOL, Ltd., Tokyo, Japan) was used to examine the capsule of S. suis following the supplier’s protocol.
Models of animal infection
Experimental infection of animals was performed in the Laboratory Animal Center of Nanjing Agricultural University with the approval of the institution’s ethics committee (Permit number SYXK (Su) 2017–0007). The zebrafish infection model detailed in previous studies was used [Citation14,Citation17,Citation31]. Exponentially grown S. suis strains were rinsed three times using phosphate-buffered saline (PBS) and adjusted to the indicated doses. Zebrafish (n = 15 per group) were received an intraperitoneal injection of 10-fold serially diluted suspensions containing 105–108 colony forming units (CFU) of bacteria. For 7 days post-infection, the mortality of the fish was recorded. The Reed and Muench method [Citation32] was used to calculate the lethal dose (LD)50. A highly virulent S. suis strain, serotype 2 meningitis isolate SC070731, was used as a positive control [Citation14,Citation33]. A BALB/c mouse infection model was also used to further evaluate the virulence of strain WUSS351. Ten mice per group were injected intraperitoneally with 3 × 108 CFU/mouse for each strain, and mouse mortality was monitored for 7 days.
Construction of the gene deletion mutants
Deletion mutant strains ΔT7SS and Δlcn351 were constructed via natural transformation using our previously published protocol [Citation34]. The upstream and downstream of deletion region fragments were obtained by PCR assay from WUSS351 genomic DNA employing primers ΔT7SS-A/B and ΔT7SS-C/D, or Δlcn351-A/B and Δlcn351-C/D. Primers spc-F/R or chl-F/R were used to amplify the spc or chl gene. Fusion PCR with primers ΔT7SS-A/D or Δlcn351-A/D was used to ligate the three amplification products. One microgram of the fusion fragment and the ComS13–21 peptide (DFSTSWWNF, 250 μM) (ChinaPeptides Co., Ltd., Shanghai, China) were mixed with 100 μL of strain WUSS351 (OD600 = 0.04–0.06) and incubated statically at 37°C for 2 h. After that, the mixture was spread on THB Spc or THB Chl agar and incubated at 37°C. Single colonies were selected and subjected to PCR to detect the deletion mutant using primers ΔT7SS-E/F or Δlcn351-E/F. Sequencing then confirmed the deletion mutant. Table S2 lists the PCR primers used.
Testing antimicrobial susceptibility
Minimum inhibitory concentrations (MICs) of the following antimicrobials toward strain WUSS351 were determined by the broth microdilution method: rifampin, β-lactam (cefotaxime, amoxicillin, and penicillin), oxazolidinone (linezolid), glycopeptide (vancomycin), fluoroquinolones (enrofloxacin), amphenicols (florfenicol and chloramphenicol), lincosamides (lincomycin and clindamycin), aminoglycosides (gentamycin and spectinomycin), macrolides (tilmicosin, tulathromycin, erythromycin, and azithromycin), tetracyclines (doxycycline), and polypeptide (bacitracin) as described in our previous study [Citation14]. The breakpoints with resistant values were determined based on our previous study [Citation14].
RNA extraction and quantitative real-time reverse transcription PCR (RT-qPCR)
The bacterial RNA was extracted using a FastRNA Pro Blue Kit (MP biomedicals, Santa Anna, CA, USA). Our previous study [Citation31] describes the detailed procedure for qRT-PCR analysis. Briefly, a reverse transcription kit (Vazyme, Nanjing, China) was used to produce cDNA from the RNA. The quantitative real-time PCR (qPCR) step, using the cDNA as the template, was carried out on a QuantStudio 6 Flex apparatus (Thermo Fisher Scientific, Shanghai, China) using a SYBR Premix Ex TaqTM (Takara) kit following the supplier’s protocols. Table S2 lists the specific primers used for RT-qPCR. The transcript level of the rpoB gene was constant under all conditions; therefore, its transcription level was used to normalize that of the target genes. The 2−ΔΔCT method was used to calculate the relative fold change. Results are shown as the mean ± the standard error of the mean (SEM) of three independent experiments.
Western blotting analysis
The strain WUSS351 in the mid-log phase was subjected to centrifugation at 8,000 × g at 4°C for 15 min. For the whole-cell proteins, PBS was used to wash the bacterial pellet three times, which was then suspended in 1× SDS-PAGE sample loading buffer and boiled for 5 min. Secreted proteins were extracted according to our previously published protocol [Citation35]. SDS-PAGE was used to separate the proteins in the samples, followed by transfer via semi-dry blotting (GE Healthcare, Chicago, IL, USA) onto PVDF membranes using a TE77 instrument (GE Healthcare). 5% skimmed milk in PBS containing 0.05% Tween-20 (PBST) was then used to block the membranes for 2 h at 37°C. Anti-FLAG antibodies recognizing Sequential Peptide Affinity (SPA)-tagged proteins [Citation36] (Sigma) or anti-GroEL antibodies [Citation35] were incubated with the membranes, followed by three washes with PBST buffer, and then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Boster, Wuhan, China). Finally, the immunoreactive proteins on the membranes were detected using an ECL Western blotting kit (Tanon, Shanghai, China) on a Tanon 5100 instrument.
Tonsillar microbiota analysis by 16S rRNA gene sequencing
Four tonsils collected from four healthy pigs were minced, weighed, and suspended at 1:9 (wt/vol) in sterile PBS. DNA was extracted directly from the tonsils for culture-independent analysis using the cetyltrimethylammonium bromide method [Citation37] and sent to Novogene Bioinformatics Technology Co., Ltd. for 16S rRNA gene sequencing. For culture-dependent analysis, tonsil samples were then processed for 80 seconds at a setting of 6.0 in a Homogenizer (MP biomedicals) to release the bacteria from the tonsillar matrix. According to a previous study [Citation10], six different media were used to cultivate a wide variety of bacteria under aerobic or anaerobic conditions. The tonsil homogenate of tonsils was added into the following six media at a ratio of 1:50 (vol/vol): Columbia broth (Hopebio, Qingdao, China), Tryptone soybean broth (TSB, Hopebio), MacConkey (Hopebio), THB, Gifu anaerobic medium (GAM, Hopebio), and Brain heart infusion (Hopebio) supplemented with 10 mg/mL each of hemin and NAD (abbreviated as BHIXV). The synthesized 73-aa Lcn351 peptide (20 μg/mL), corresponding to the putative mature form of Lcn351, (AVQYPDGGVWTYGASNGGAFSNYYHGKKEHSSTVVSRKDSRSAKGSAGPGQTSKAYIKTSFGEPAAFYYDFWQ) (ChinaPeptides Co., Ltd.) was added into the above media as the Lcn351 groups, and for the control group, an equal volume of H2O was added. The media with the tonsil homogenate, including Columbia broth, TSB, MacConkey, THB, and BHIXV, were incubated aerobically at 37°C for 8 h with 5% CO2, and the GAM medium with homogenate was incubated anaerobically for 8 h at 37°C. Bacteria were collected by centrifugation at 8,000 × g for 10 min, washed two times with PBS, resuspended, and mixed together from the six media for each tonsil. The DNA extracted from mixed bacteria was sent to Novogene Bioinformatics Technology Co., Ltd. for 16S rRNA gene sequencing. Sequences reads were grouped into operational taxonomic units (OTUs) at a sequence similarity level of ≥97% for a taxonomic assignment using Uparse software (http://drive5.com/uparse/) [Citation38,Citation39], and species annotation was conducted through the SSUrRNA Database at the Silva Database (https://www.arb-silva.de/). Subsequent alpha and beta diversity analyses were performed based on the normalized data. The relative abundance of OTUs was represented as the mean ± SEM.
Culture with supernatant or the Lcn351 peptide
To culture different bacterial targets in the presence of strain WUSS351 or Δlcn351 supernatants, the supernatants of overnight (10 h) culture (strains WUSS351 or Δlcn351, 25 mL) were centrifuged at 13,000 × g. The supernatants were filtered through a 0.22 mm pore size filter (BIOFIL, Indore, India). Bacterial targets (107 cells) were inoculated at 37°C into 2.5 mL of strain WUSS351 or Δlcn351 supernatants and 2.5 mL of fresh medium: nutrient broth (Hopebio) for Bacillus subtilis; MRS broth (Hopebio) for Lactobacillus lactis and Lactobacillus rhamnosus; LB for E. coli; THB for S. suis, S. dysgalactiae, Streptococcus equi subsp. zooepidemicus, Streptococcus agalactiae, Aeromonas hydrophila, and Klebsiella pneumoniae. At 6 h after inoculation, the cultures were serially diluted and plated on the appropriate agar plates. For in vitro assays, 107 bacterial cells were inoculated into different concentrations of the synthesized Lcn351 peptide (10 or 20 μg/mL) into 5 mL of the medium at 37°C as test groups. For the control groups, an equal volume of H2O was added. At 6 h after inoculation, the cultures were serially diluted and plated. The results were calculated as the relative growth rate (RGR) = AGR Lcn351 or H2O /AGR Lcn351, where AGR represents the absolute growth rate = CFU final /CFU initial. Data are expressed as the mean ± SEM.
Bacterial contact-dependent competition experiments
Similar to the rifampin (Rif)-resistant induced strain S. suis P1/7-Rif [Citation40], L. lactis strain WUQT017, and S. dysgalactiae strain WUQT020 were selected using increasing amounts of antibiotics to produce a Rif-resistant phenotype. The stepwise induction method was used to produce nalidixic acid (Nal)-resistant phenotypes of A. hydrophila strain WUQT018, K. pneumoniae strain WUQT019, and E. coli strain WUQT022. For the competition assays, the attacker and prey strains were diluted to an initial OD600 of 0.6 using THB. The attacker and prey cells were mixed at a ratio of 10:1 (CFU attacker : CFU prey), centrifuged, and resuspended in 50 μL of PBS. Two microliters of the mixture were spotted on LB agar medium and grown for different times at 37°C with 5% CO2 for Gram-positive bacteria. The mixture was spotted onto a nitrocellulose membrane on LB agar medium for the competition experiments for Gram-negative bacteria. The CFU of the prey and attacker strains was calculated to determine the starting ratio. The competition was determined by excising the agar with cell growth and resuspending the cells in 1 mL PBS. The final count of the prey was calculated by plating serial dilutions onto appropriate agar plates with selective antibiotics. The information related to the antibiotics and agar plates for the prey was as follows: S. suis strain P1/7-Rif and S. dysgalactiae strain WUQT020-Rif, 8 μg/mL Rif and THB agar plates; S. suis strain ΔT7SS, 100 μg/mL Spc and THB agar plates; L. lactis strain WUQT017-Rif, 8 μg/mL Rif and MRS agar plates; A. hydrophila strain WUQT018-Nal, 32 μg/mL Nal and TSB agar plates; K. pneumoniae strain WUQT019-Nal, 32 μg/mL Nal and MacConkey agar plates; E. coli strain WUQT022-Nal, 32 μg/mL Nal and LB agar plates.
Statistical analysis
The Log-rank (Mantel-Cox) test was used to analyze the survival curve in the animal experiments. A two-tailed paired t test was used to compare the Lcn351 and control groups. A two-tailed unpaired t test was used for the rest of the experiments. GraphPad Prism software was used to perform all the statistical analyses.
Results
WUSS351 belongs to the NCL4 type and is an encapsulated and virulent strain
Strain WUSS351 has a genome comprising a single circular chromosome of 2,283,286 bp, but no plasmids. Table S3 summarizes the general genomic features. Table S4 lists the 39 predicted genomic islands (GIs). To determine the cps genotype of strain WUSS351, phylogenetic tree analysis based on the sequence of the cps cluster in all S. suis strains (29 serotypes and 28 NCL types) was performed, which showed that strain WUSS351 is located at Group 3, together with NCL4 strain YS73, NCL5 strain YS35, serotype 9 strain 22083, and NCL26 strain NJ1112 (). The sequence of cps cluster of strain WUSS351 shared 98.86% nucleotide identity with that of NCL4 strain YS73 (GenBank: KM972278.1) (), confirming that strain WUSS351 belongs to the NCL4 type. Transmission electron microscopy (TEM) analysis showed that strains WUSS351 and SC070731 (as a positive control) [Citation6] were encapsulated ().
Figure 1. The capsule and pathogenicity of strain WUSS351. (a) The phylogenetic tree of S. suis cps clusters from different serotypes or genotypes, created using the Neighbor-Joining method. (b) The comparative analysis of cps gene clusters between S. suis NCL4 strains WUSS351 and YS73. (c) The capsule of S. suis strains WUSS351 and SC070731 (an encapsulated strain control), as observed using TEM. (d) The survival curve of mice injected with strains WUSS351 and SC070731 (a virulent strain control) at the dose of 3 × 108 CFU/mouse or PBS (a negative control). Asterisks indicate pairs of significantly different values (n = 10, the Log-rank (Mantel-Cox) test, **** indicates p < 0.0001, ns indicates not significant).
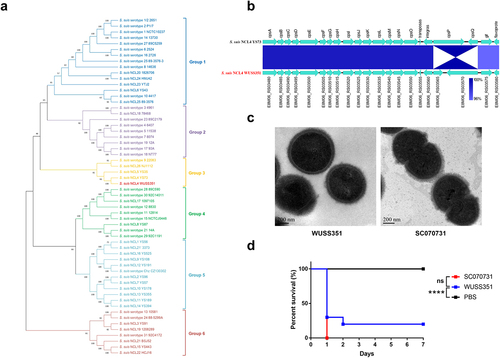
The zebrafish infection model was first used to evaluate the virulence of strain WUSS351. The LD50 value of strain WUSS351 was 1.1 × 106 CFU/fish, similar to that of the virulent control strain SC070731 [Citation14,Citation33] (8.6 × 105 CFU/fish). Then, their virulence was further determined in a mouse infection model. As shown in , strains WUSS351 and SC070731 induced 80% and 100% mortality, respectively, and a Log-rank (Mantel-Cox) test showed that their virulence was not significantly different.
Strain WUSS351 is multidrug-resistant
To determine which antimicrobials strain WUSS351 is susceptible to, a MIC assay was performed for 19 antimicrobials comprising 11 classes. WUSS351 showed resistance to nine antimicrobials of five classes (). A strain that is resistant to ≥ 3 classes of antimicrobials is considered multidrug-resistant [Citation41]. Thus, strain WUSS351 is multidrug-resistant. A BLAST search for acquired antimicrobial resistance genes in strain WUSS351 was performed to clarify the antibiotic resistance mechanisms. In addition to the macrolide-lincosamide-streptogramin B resistance gene erm(B) and the tetracycline resistance gene tet(O), other antimicrobial resistance genes were found, including an ABC transporter system gene sstFEG and Bce system genes bceBA and bceSR, which confer bacitracin resistance in S. suis [Citation42]. The genes erm(B) and tet(O) were in predicted GIs 19 and 20, respectively (), while the bacitracin resistance genes sstFEG, bceBA, and bceSR were in an integrative and conjugative element (ICE) (ICEWUSS351) (Figure S1). We observed a chromosomal mutation of pbp2x, which is involved in resistance to penicillin [Citation43]. Thus, the resistance phenotype is in accordance with the genotype of antimicrobial resistance.
Table 1. MICs of strain WUSS351
Strain WUSS351 harbors a novel T7SS that mediates contact-dependent antagonism of S. suis
To investigate the phylogenetic relationship between strain WUSS351 and 51 strains whose complete genomic sequences are available from NCBI, phylogenetic tree analysis based on whole genomic sequences was performed. As shown in , strain WUSS351 was closely related to S. suis serotype 4 strain HA1003, serotype Chz strain 0061, serotype 31 strain 1081, serotype 9 strain 9401240, and serotype 5 strain HN105, and distantly related to serotype 2 strains SC19, SC84, ZY05719, 05HAH33, SS2–1, 98HAH33, S10, P1/7, BM407, S735, GZ1, and SC070731.
Figure 2. Comparative genome analysis. (a) The genetic distance among S. suis strains was calculated using the OrthoANIu algorithm. Based on the distance data, Neighbor-Joining method was used to construct the phylogenetic tree. S. pneumoniae strain TIGR4 was used as an out-group. (b) The Mauve software was used to align 52 S. suis genomes, and the Circos software was used to display comparison results. To better present the comparative genome analysis results, strain WUSS351 and representative strains from different branches of the phylogenetic tree shown in Figure 2a were selected. A 25 kb genomic island was predicted in the genome of strain WUSS351 but not in the other 51 S. suis strains.
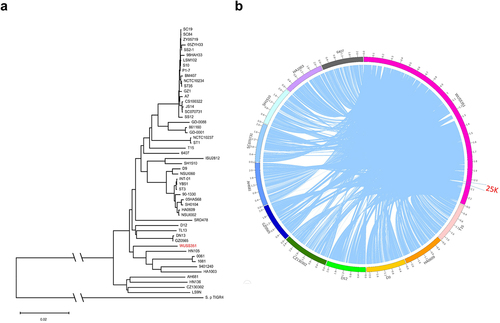
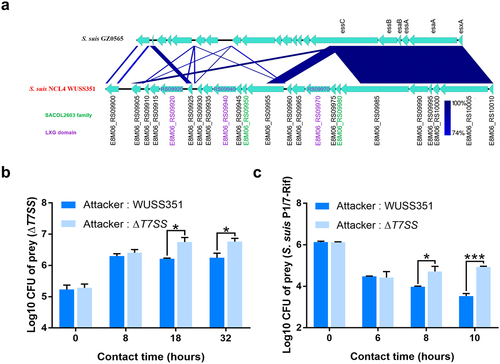
Comparative genomic analysis showed that strain WUSS351 contains a 25-kb GI that was not detected in the other 51 S. suis strains (). The 25-kb GI contains a novel T7SS cluster (), and several S. suis strains harbor a T7SS with 40–45% sequence similarity to this novel T7SS (Table S5). In Gram-positive bacteria, the T7SS mediates contact-dependent antagonism during bacterial competition [Citation44], which motivated us to investigate the capacity of this novel T7SS to mediate inter-bacterial antagonism. As shown in , significant contact-dependent antagonism of ΔT7SS was observed in co-cultured with wild-type strain WUSS351 at 18 h and 32 h, compared with co-cultured with the strain lacking T7SS. A similar contact-dependent antagonism of S. suis virulent reference strain P1/7-Rif was observed when co-cultured with wild-type strain WUSS351 at 8 h and 10 h, compared with co-culture with the strain lacking the T7SS (). The wild-type strain and ΔT7SS showed no significant changes in growth in THB medium (Figure S2). However, the T7SS of WUSS351 did not exhibit its capacity of inter-bacterial antagonism when interacting with other five strains isolated from pig tonsils using six different media described in tonsillar microbiota analysis, consisting of Lactococcus lactis strain WUQT017-Rif, Aeromonas hydrophila strain WUQT018-Nal, Klebsiella pneumoniae strain WUQT019-Nal, Streptococcus dysgalactiae strain WUQT020-Rif, and Escherichia coli strain WUQT022-Nal (data not shown).
Figure 3. The T7SS of strain WUSS351. (a) Comparison of the sequence of the T7SS clusters between strains WUSS351 and GZ0565. (b) Results of competition experiments performed with WUSS351 or ΔT7SS as attacker strains and ΔT7SS as the prey strain. The number of the final prey cells was determined at 8, 18, and 32 h by serially diluting cultures onto LB+Spc agar plates. (c) Results of competition experiments performed with WUSS351 or ΔT7SS as attacker strains and S. suis strain P1/7-Rif as the prey strain. The number of the final prey cells was determined at 6, 8, and 10 h by serially diluting cultures onto LB+Rif agar plates. The data in the graphs represent the means ± SEM, and asterisks indicate pairs of significantly different values (n = 3, *** indicates p <0.001, * indicates p <0.05, two-tailed unpaired t test).
Lactococcin Lcn351 inhibits the growth of Bacillus subtilis and S. suis
Further genomic analysis found that strain WUSS351 contains two different bacteriocins. One is a lantibiotic bacteriocin (encoded by E8M06_RS05390). The nucleotide sequence of this bacteriocin locus in strain WUSS351 shares 100% identity with that of suicin3908 locus in S. suis 3908 [Citation45] (Figure S3a). Western blotting analysis showed that strain WUSS351 could express this bacteriocin as a SPA-tagged fusion protein (Figure S3b). Vaillancourt et al. reported that Suicin3908 was resistant to high temperature, pH, and proteases digestion and was active against S. suis and Staphylococcus hyicus [Citation45].
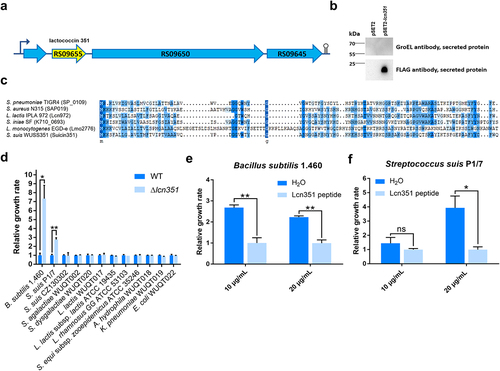
The characteristics of Suicin3908 have been studied; therefore, we focused on the second bacteriocin in strain WUSS351, a lactococcin972-like bacteriocin (named as Lcn351, encoded by E8M06_RS09655) (). Western blotting analysis confirmed that strain WUSS351 could secrete Lcn351 as a SPA-tagged fusion protein (). As a control, GroEL, a cytoplasmic protein in S. suis [Citation35], was not detected among the secreted proteins, reflecting no impurities in the secreted proteins from autolytic products. As shown in , the amino acid sequence of Lcn351 shares 37.63% identity with Lcn972 of L. lactis strain IPLA 972, 29.85% identity with Lmo2776 of Listeria monocytogenes strain EGD-e, 28.12% identity with SP_0109 of Streptococcus pneumoniae strain TIGR4, 35.42% identity with SAP019 of Staphylococcus aureus strain N315, and 40.62% identity with K710_0693 of Streptococcus iniae strain SF. The mRNA expression level of genes in lcn351 locus was significantly higher in the stationary phase than in the exponential phase in THB (Figure S4). To determine the inhibitory spectrum of Lcn351, 12 strains were selected, consisting of six strains isolated from pig tonsils, three strains isolated from diseased pigs, and three strains that could be targets of the lactococcin972-like bacteriocins according to previous reports [Citation23,Citation46]. After incubating with the supernatant of wild-type (WT) or Δlcn351 from the stationary phase for 6 h, the growth of B. subtilis strain 1.460 and S. suis strain P1/7 was dramatically inhibited in the WT supernatant group compared with that in the Δlcn351 supernatant group (). However, Lcn351 did not affect the growth of the other ten strains. To further confirm the function of Lcn351, the activity of its synthesized peptide was analyzed on B. subtilis strain 1.460 and S. suis strain P1/7. Synthesized Lcn351 was observed to inhibit the growth of these two strains (, f).
Figure 4. Lcn351 targets B. subtilis and S. suis. (a) The genetic organization of the lactococcin cluster (blue) and Lcn351 (yellow) in strain WUSS351. (b) The secreted proteins extracted from strain WUSS351 expressing Lcn351 fused with a SPA tag from its own promoter of the cluster (pSET2-lcn351) or control plasmid pSET2 were probed with FLAG or GroEL antibodies. (c) Alignment of the amino acid sequences of Lcn972 family bacteriocins: SP_0109 of S. pneumoniae strain TIGR4, SAP019 of S. aureus strain N315, Lcn972 of L. lactis strain IPLA 972, K710_0693 of S. iniae strain SF, and Lmo2776 of L. monocytogenes strain EGD-e. (d) The relative growth rate of different bacteria calculated after 6 h of incubation with supernatants of WT or Δlcn351. The relative growth rate of B. subtilis 1.460 (e) and S. suis P1/7 (f) after 6 h of incubation in the presence of Lcn351 peptide (10 or 20 mg/mL) or H2O; the relative growth rate was compared with Lcn351 group. The graph shows the means ± SEM; asterisks indicate pairs of significantly different values (n = 3, ** indicates p <0.01, * indicates p <0.05, ns indicates not significant, two-tailed unpaired t test).
Lactococcin Lcn351 mainly targets S. suis in pig tonsils
To determine the target of Lcn351 in pig tonsils, the microbiota composition of pig tonsils was first verified by 16S rRNA gene sequencing. We found members of 15 phyla, 18 classes, 38 orders, 63 families, 122 genera, and 91 species of bacteria in one or more tonsils (Table S6) through sequencing the culture-independent tonsillar microbiome (). We found 12 of 15 phyla, 11 of 18 classes, 22 of 38 orders, 31 of 63 families, and 40 of 122 genera were also reported as pig tonsillar microbiota in a previous report [Citation47]. The detailed list of pig tonsillar microbiota reported in both studies is summarized in Table S6. We defined the core microbiome of the pig tonsils as those presented in two or more samples and had a relative abundance exceeding 1% (Tables S6 and S7). Notably, the average relative abundance of Streptococcus genus was 14.13%, marking it as one member of the core microbiomes in pig tonsils in all tonsils (Table S6). Distribution at the species level followed well from the genus level data. S. suis was one of the dominant microbial communities in porcine tonsils, with an average relative abundance of 13.52% (Table S6), consistent with the previous study wherein S. suis was present in most pig tonsils [Citation47].
Figure 5. Lcn351 targets S. suis in pig tonsils microbiota. (a) A schematic diagram of the culture-independent and culture-dependent pig tonsillar microbiome analysis. Please refer to the Materials and methods section for the detailed protocols. (b) The relative abundance of species in the culture-independent and culture-dependent communities of pig tonsils. (c) The relative abundance of S. suis in the culture-dependent community of pig tonsils. The data in the graphs represent the means ± SEM, and asterisks indicate pairs of significantly different values (n = 4, * indicates p < 0.05, two-tailed unpaired t test).
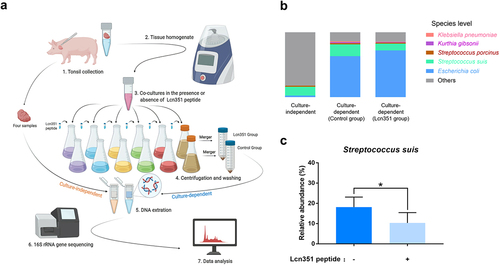
Next, we compared the microbiota compositions cultured from pig tonsils with or without synthesized Lcn351 using 16S rRNA gene sequencing. According to a previous study [Citation10], as shown in , we used six different media to cultivate various bacteria from pig tonsils under aerobic or anaerobic conditions. The cultured bacteria from each group were merged for 16S rRNA gene sequencing. As expected, the microbiota composition by culture-dependent analysis in the control group (without synthesized Lcn351) was significantly different from that obtained by culture-independent analysis (Figure S5a, b). The complete list of all taxons identified in the culture-dependent microbiome in the control group is summarized in Table S8. The most significant differences in microbiota composition included increased levels of Enterobacteriaceae (from 0.29% relative abundance by culture-independent analysis to 65.20% relative abundance by culture-dependent analysis) and Staphylococcaceae (from 0.42% relative abundance by culture-independent analysis to 9.13% relative abundance by culture-dependent analysis) (Figure S5a, b, and Tables S6 and S8). As shown in , compared with the culture-independent microbial community, S. suis was still one of the dominant bacteria in the culture-dependent microbial community, although E. coli became the most dominant bacteria. The culture-dependent microbiota composition of pig tonsils was dramatically altered after the addition of Lcn351. Species whose relative abundance exceeded 1% by culture-dependent analysis in the control group are presented in . The genera whose relative abundance exceeded 1% by culture-dependent analysis in the control group are presented in Figure S6a. The complete list of all taxons identified in the culture-dependent microbiome in the Lcn351 group is summarized in Table S9. Comparing the relative abundance at the genus level shown in Tables S8 and S9, the relative abundance of the Streptococcus genus was reduced from 23.08% to 13.15% after adding Lcn351, while that of the Escherichia genus increased from 63.22% to 72.13%. The reduced relative abundance of Streptococcus genus was mainly caused by the decreased relative abundance of S. suis (from 18.15% to 10.32%) (). In addition, the relative abundances of S. porcinus, K. pneumoniae, and Kurthia gibsonii also decreased in the group with Lcn351; however, their changes were not significant (Figure S6b-d). These findings indicated that S. suis is the primary target of Lcn351 in pig tonsils.
Discussion
To date, the only available information for S. suis NCL4 was that it could be isolated from healthy pigs [Citation7,Citation48] and diseased pigs [Citation8]. In this study, we found that S. suis NCL4 strain WUSS351 isolated from a healthy pig tonsil is virulent and has several systems that promote its niche competition in pig tonsils. Strain WUSS351 is multidrug-resistant, which might confer a competitive advantage to preferentially colonize the host over other microbiota constituents in the presence of antimicrobial selective pressures, leading to an increased prevalence. In addition, some bacteria from the host microflora can produce bacitracin [Citation42]. Thus, strain WUSS351’s resistance to bacitracin might represent a competitive advantage in the host environment. Our previous study found that sstFEG is present mainly in S. suis virulent strains isolated from pigs or humans [Citation42]. sstFEG not only confers resistance to bacitracin but also contributes to S. suis virulence via a competitive survival advantage in vivo [Citation42].
T7SS is pivotal for pathogenic bacterial competitive capability and colonization. S. aureus utilizes its T7SS to secrete toxins that inhibit the growth of the rival bacteria, such as EsaD, causing DNA damage as a nuclease toxin [Citation49], and TspA, targeting the periplasm of competitor bacteria as a membrane-depolarizing toxin [Citation50]. The T7SS of Streptococcus gallolyticus subsp. gallolyticus contributes to its murine gut colonization by enhancing adherence capacity [Citation21]. We first identified the T7SS in S. suis virulent strain GZ0565 and demonstrated that its substrate, EsxA, contributed to S. suis virulence [Citation35]. However, the sequence coverage of this novel T7SS between strains WUSS351 and GZ0565 is only 42%. After comparing the T7SS cluster sequence with that of serotype 9 virulent strain GZ0565, strain WUSS351 was observed to harbor three LXG domain-containing proteins (with 34–58% identity, encoded by E8M06_RS09920, E8M06_RS09940, and E8M06_RS09970) in the T7SS (). The LXG domain is present in the N-terminus of a group of bacterial toxins predicted to be exported by the T7SS [Citation51]. In addition to the N-terminal LXG domain, LXG family toxins usually contain a C-terminal variable toxin domain [Citation51]. The three LXG-containing effectors of Streptococcus intermedius T7SS mediated inter-bacterial antagonism, and one of them degrades lipid II required for cell wall biosynthesis [Citation52]. Additionally, the SACOL2603 family proteins are also considered as potential T7SS effectors and are similar in length and sequence to WXG family proteins [Citation53]. Two SACOL2603 family proteins (encoded by E8M06_RS09950 and E8M06_RS09980) were predicted (). Thus, it would be interesting to determine their functions in both virulence and bacterial competition in the future.
Many bacterial species produce small antimicrobial peptides termed bacteriocins, which are believed to function in inter-bacterial competition. The bacteriocin BlpC of S. pneumoniae contributes to competition and fitness by targeting neighboring bacteria for lysis [Citation54]. The bacteriocin Nukacin IVK45 increases the competitive capability of Staphylococcus epidermidis in human nasal microbiotas [Citation55]. L. monocytogenes bacteriocin LLS contributes to virulence and intestinal survival in a murine oral infection model, inhibits the growth of L. monocytogenes, L. lactis, and S. aureus, and promotes persistence in murine intestinal microbiota [Citation56]. We found that strain WUSS351 possesses two bacteriocins. One is a lantibiotic bacteriocin whose nucleotide shares 100% identity with suicin3908 of S. suis strain 3908. Suicin3908 has membrane permeabilization activity and shows antibacterial activity against ten pathogenic S. suis strains (members of the ST1, ST25, or ST28 groups) and S. hyicus [Citation45], indicating that this bacteriocin might provide an inter-bacterial competitive advantage. The other bacteriocin is Lcn351, belonging to the lactococcin 972 family. Lactococcin Lcn972 of L. lactis subsp. lactis specifically kills other lactococci by inhibiting bacterial cell division via blocking the incorporation of cell wall precursors in the septum [Citation46]. Lactococcin Lmo2776 of L. monocytogenes mainly targets the intestinal bacteria Prevotella copri to reduce its abundance and avoid the excessive inflammation caused by P. copri, which affects L. monocytogenes colonization in the host intestine [Citation57]. Lactococcin Sil of S. iniae targets B. subtilis, inhibits host cell innate immune responses and increases bacterial infection by promoting dissemination and colonization [Citation23]. Our study found lactococcin972-like bacteriocin Lcn351 mainly targeted S. suis based on the following reasons: 1) in antibacterial activity analysis, 12 strains mainly isolated from pig tonsils or diseased pigs were selected; in addition to B. subtilis strain 1.460 reported as a target of lactococcin972-like bacteriocins [Citation23,Citation57], Lcn351 only inhibited the growth of S. suis strain P1/7 (), not the other ten strains; 2) by 16S rRNA gene sequencing of the culture-dependent pig tonsillar microbiome, more than 22 species were identified; the significant change of the relative abundance after the addition of Lcn351 was only observed in S. suis (Tables S8 and S9), not the other species. We observed that Lcn351 inhibited the growth of S. suis in the tonsil. However, only two S. suis strains (serotype 2 strain P1/7 and serotype Chz strain CZ130302) were used in antibacterial activity analysis. Therefore, we did not know if this inhibition effect of Lcn351 was limited to a specific type or serotype of S. suis.
Inter-bacterial competition in the porcine tonsil during co-colonization might have important implications for the epidemiology of S. suis, potentially impacting serotype distributions, strain prevalence, and disease progression. Efficient removal of competitors could allow S. suis strain WUSS351 to colonize the tonsils more efficiently and for longer periods, which would increase its transmission potential. Currently, we are studying the correlation between the antimicrobial systems and S. suis’ ability to colonize and induce disease in a larger panel of clinical isolates.
Supplemental Material
Download Zip (1.8 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete genome sequence of strain WUSS351 was deposited in GenBank and received the accession number NZ_CP039462.1. The data supporting this study’s findings are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Goyette-Desjardins G, Auger JP, Xu J, et al. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect. 2014;3(6):e45. DOI:10.1038/emi.2014.45
- Ferrando ML, de Greeff A, van Rooijen WJ, et al. Host-pathogen interaction at the intestinal Mucosa correlates with zoonotic potential of Streptococcus suis. J Infect Dis. 2015;212(1):95–105.
- Segura M, Fittipaldi N, Calzas C, et al. Critical Streptococcus suis virulence factors: are they all really critical? Trends Microbiol. 2017;25(7):585–599. DOI:10.1016/j.tim.2017.02.005
- Huang J, Liang Y, Guo D, et al. Comparative genomic analysis of the ICESa2603 family ICEs and spread of erm(b)- and tet(o)-carrying transferable 89K-subtype ICEs in Swine and Bovine Isolates in China. Front Microbiol. 2016;7:55.
- Pan Z, Ma J, Dong W, et al. Novel variant serotype of Streptococcus suis isolated from piglets with meningitis. Appl Environ Microbiol. 2015;81(3):976–985.
- Huang J, Liu X, Chen H, et al. Identification of six novel capsular polysaccharide loci (NCL) from Streptococcus suis multidrug resistant non-typeable strains and the pathogenic characteristic of strains carrying new NCLs. Transbound Emerg Dis. 2019;66(2):995–1003. DOI:10.1111/tbed.13123
- Zheng H, Ji S, Liu Z, et al. Eight novel capsular polysaccharide synthesis gene loci identified in nontypeable Streptococcus suis isolates. Appl Environ Microbiol. 2015;81(12):4111–4119.
- Zheng H, Qiu X, Roy D, et al. Genotyping and investigating capsular polysaccharide synthesis gene loci of non-serotypeable Streptococcus suis isolated from diseased pigs in Canada. Vet Res. 2017;48(1):10.
- Bojarska A, Janas K, Pejsak Z, et al. Diversity of serotypes and new cps loci variants among Streptococcus suis isolates from pigs in Poland and Belarus. Vet Microbiol. 2020;240:108534.
- Lowe BA, Marsh TL, Isaacs-Cosgrove N, et al. Microbial communities in the tonsils of healthy pigs. Vet Microbiol. 2011;147(3–4):346–357.
- Vötsch D, Willenborg M, Weldearegay YB, et al. Streptococcus suis - the “Two Faces” of a pathobiont in the porcine respiratory tract. Front Microbiol. 2018;9:480.
- Arndt ER, Farzan A, Slavic D, et al. An epidemiological study of Streptococcus suis serotypes of pigs in Ontario determined by a multiplex polymerase chain reaction. Can Vet J. 2018;59(9):997–1000
- Liang P, Wang M, Gottschalk M, et al. Genomic and pathogenic investigations of Streptococcus suis serotype 7 population derived from a human patient and pigs. Emerg Microbes Infect. 2021;10(1):1960–1974. DOI:10.1080/22221751.2021.1988725
- Wang X, Sun J, Bian C, et al. The population structure, antimicrobial resistance, and pathogenicity of Streptococcus suis cps31. Vet Microbiol. 2021;259:109149. DOI:10.1016/j.vetmic.2021.109149
- Swildens B, Nielen M, Wisselink HJ, et al. Elimination of strains of Streptococcus suis serotype 2 from the tonsils of carrier sows by combined medication and vaccination. Vet Rec. 2007;160(18):619–621.
- Ngo TH, Tran TB, Tran TT, et al. Slaughterhouse pigs are a major reservoir of Streptococcus suis serotype 2 capable of causing human infection in southern Vietnam. PLoS One. 2011;6(3):e17943.
- Dong X, Chao Y, Zhou Y, et al. The global emergence of a novel Streptococcus suis clade associated with human infections. EMBO Mol Med. 2021;13(7) ;e13810. doi:10.15252/emmm.202013810.
- Bottai D, Groschel MI, Brosch R. Type VII secretion systems in gram-positive bacteria. Curr Top Microbiol Immunol. 2017;404:235–265.
- Houben D, Demangel C, van Ingen J, et al. ESX-1-Mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14(8):1287–1298. DOI:10.1111/j.1462-5822.2012.01799.x
- Burts ML, Williams WA, DeBord K, et al. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. P Natl Acad Sci USA. 2005;102(4):1169–1174.
- Taylor JC, Gao XS, Xu J, et al. A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog. 2021;17(1):e1009182. DOI:10.1371/journal.ppat.1009182
- Liu Y, Shu X, Chen L, et al. The beneficial rhizobacterium Bacillus velezensis acquires iron from roots via a type VII secretion system for colonization. bioRxiv. 2021 https://doi.org/10.1101/2021.08.01.454677.
- Li MF, Zhang BC, Li J, et al. Sil: a Streptococcus iniae bacteriocin with dual role as an antimicrobial and an immunomodulator that inhibits innate immune response and promotes S. iniae infection. PLoS One. 2014;9(4): e96222. doi:10.1371/journal.pone.0096222.
- Heilbronner S, Krismer B, Brotz-Oesterhelt H, et al. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol. 2021;19(11):726–739. DOI:10.1038/s41579-021-00569-w
- Li R, Li Y, Kristiansen K, et al. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714.
- Bertelli C, Laird MR, Williams KP, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–W35.
- Yoon SH, Ha SM, Lim J, et al. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110(10):1281–1286. DOI:10.1007/s10482-017-0844-4
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549.
- Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147.
- Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. DOI:10.1101/gr.092759.109
- Wu Z, Wu C, Shao J, et al. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. Rna. 2014;20(6):882–898. DOI:10.1261/rna.041822.113
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497.
- Wu ZF, Wang WX, Tang M, et al. Comparative genomic analysis shows that Streptococcus suis meningitis isolate SC070731 contains a unique 105 K genomic island. Gene. 2014;535(2):156–164.
- Zhu Y, Dong W, Ma J, et al. Utilization of the ComRS system for the rapid markerless deletion of chromosomal genes in Streptococcus suis. Future Microbiol. 2019;14(3):207–222. DOI:10.2217/fmb-2018-0279
- Lai LY, Dai J, Tang HY, et al. Streptococcus suis serotype 9 strain GZ0565 contains a type VII secretion system putative substrate EsxA that contributes to bacterial virulence and a vanZ-like gene that confers resistance to teicoplanin and dalbavancin in Streptococcus agalactiae. Vet Microbiol. 2017;205:26–33.
- Zeghouf M, Li J, Butland G, et al. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res. 2004;3(3):463–468. DOI:10.1021/pr034084x
- Xu J, Dai W, Liang Q, et al. The microbiomes of adenoid and middle ear in children with otitis media with effusion and hypertrophy from a tertiary hospital in China. Int J Pediatr Otorhinolaryngol. 2020;134:110058.
- Tindall BJ, Rossello-Mora R, Busse HJ, et al. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60(Pt 1):249–266. DOI:10.1099/ijs.0.016949-0
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013 Oct;10(10):996–998.
- Huang J, Ma J, Shang K, et al. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: a probable mobile genetic elements reservoir for other streptococci. Front Cell Infect Microbiol. 2016;6:118.
- Schwarz S, Silley P, Simjee S, et al. Editorial: assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother. 2010;65(4):601–604. DOI:10.1093/jac/dkq037
- Ma JL, Liu J, Zhang Y, et al. Bacitracin resistance and enhanced virulence of Streptococcus suis via a novel efflux pump. BMC Vet Res. 2019;15(1):377. doi:10.1186/s12917-019-2115-2.
- Huang J, Sun J, Wu Y, et al. Identification and pathogenicity of an XDR Streptococcus suis isolate that harbours the phenicol-oxazolidinone resistance genes optrA and cfr, and the bacitracin resistance locus bcrABDR. Int J Antimicrob Agents. 2019;54(1):43–48. DOI:10.1016/j.ijantimicag.2019.04.003
- Sibinelli-Sousa S, Hespanhol JT, Bayer-Santos E. Targeting the Achilles’ heel of bacteria: different mechanisms to break down the peptidoglycan cell wall during bacterial warfare. J Bacteriol. 2021;203(7). DOI:10.1128/JB.00478-20
- Vaillancourt K, LeBel G, Frenette M, et al. Suicin 3908, a new lantibiotic produced by a strain of Streptococcus suis serotype 2 isolated from a healthy carrier pig. PLoS One. 2015;10(2):e0117245. DOI:10.1371/journal.pone.0117245
- Martinez B, Rodriguez A, Suarez JE. Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiol-Uk. 2000 Apr;146(4):949–955.
- Lowe BA, Marsh TL, Isaacs-Cosgrove N, et al. Defining the “core microbiome” of the microbial communities in the tonsils of healthy pigs. BMC Microbiol. 2012;12(1):20.
- Qiu XT, Bai XM, Lan RT, et al. Novel capsular polysaccharide loci and new diagnostic tools for high-throughput capsular gene typing in Streptococcus suis. Appl Environ Microb. 2016;82(24):7102–7112
- Cao ZP, Casabona MG, Kneuper H, et al. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. 2017;2(1). DOI:10.1038/nmicrobiol.2016.183
- Ulhuq FR, Gomes MC, Duggan GM, et al. A membrane-depolarizing toxin substrate of the Staphylococcus aureus type VII secretion system mediates intraspecies competition. Proc Natl Acad Sci U S a. 2020;117(34):20836–20847.
- Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011 Jun;39(11):4532–4552.
- Whitney JC, Peterson SB, Kim J, et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife. 2017;6. DOI:10.7554/eLife.26938
- Warne B, Harkins CP, Harris SR, et al. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics. 2016;17. DOI:10.1186/s12864-016-2426-7
- Wholey WY, Kochan TJ, Storck DN, et al. Coordinated bacteriocin expression and competence in streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog. 2016;12(2):e1005413. DOI:10.1371/journal.ppat.1005413
- Janek D, Zipperer A, Kulik A, et al. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog. 2016;12(8):e1005812. DOI:10.1371/journal.ppat.1005812
- Quereda JJ, Dussurget O, Nahori MA, et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. P Natl Acad Sci USA. 2016;113(20):5706–5711.
- Rolhion N, Chassaing B, Nahori M-A, et al. A Listeria monocytogenes bacteriocin can target the commensal Prevotella copri and modulate intestinal infection. Cell Host Microbe. 2019;26(5):691–701. e5. DOI:10.1016/j.chom.2019.10.016
