ABSTRACT
The role of secretion chaperone-regulated virulence proteins in the pathogenesis of infective endocarditis (IE) induced by viridans streptococci such as Streptococcus mutans is unclear. In this study, we investigated the contribution of the foldase protein PrsA, a putative parvulin-type peptidyl-prolyl isomerase, to the pathogenesis of S. mutans-induced IE. We found that a prsA-deficient strain had reduced virulence in terms of formation of vegetation on damaged heart valves, as well as reduced autolysis activity, eDNA release and biofilm formation capacity. The secretion and surface exposure of AtlA in vitro was reduced in the prsA-deficient mutant strain, and complementation of recombinant AtlA in the culture medium restored a wild type biofilm phenotype of the prsA-deficient mutant strain. This result suggests that secretion and surface localization of AtlA is regulated by PrsA during biofilm formation. Together, these results demonstrate that S. mutans PrsA could regulate AtlA-mediated eDNA release to contribute to biofilm formation in the pathogenesis of IE.
Introduction
Bacterial biofilm formation is a determining virulence factor for bacteria that cause infectious disease [Citation1]. Biofilms not only mediate bacterial colonization but also contribute to bacterial resistance to antibiotic treatment [Citation2,Citation3]. Infective endocarditis (IE) is a typical infectious disease related to formation of biofilms that are frequently induced by staphylococci or oral streptococci [Citation1,Citation4]. In contrast to formation of biofilms on polystyrene surfaces in vitro that mainly comprise a carbohydrate matrix, our previous studies demonstrated that host factors, platelets, and neutrophil extracellular traps (NETs) contribute to bacteria biofilm formation and vegetation maturation in the pathogenesis of IE [Citation5,Citation6]. In addition, bacterial extracellular DNA (eDNA) plays a crucial role in Streptococcus mutans biofilm formation inside vegetation on affected heart valves [Citation7,Citation8].
Bacterial eDNA release to biofilm matrices is mainly mediated by two specific mechanisms: autolysis and vesicle-associated active secretion [Citation9]. Bacterial autolysin mediates bacterial lysis with consequent release of eDNA into biofilms formed by diverse species [Citation10,Citation11]. Our previous studies demonstrated that AtlA, an S. mutans autolysin, mediates bacterial eDNA release, which contributes to bacterial biofilm formation on heart valves in IE [Citation7]. AtlA does not contain the typical LPXTG cell wall anchor domain [Citation12]. As such, the mechanism of AtlA surface localization is unclear. A recent study indicated that AtlA specifically binds to immature serotype c carbohydrates in the cell wall and mediates bacterial cell wall separation [Citation13]. These data implied that AtlA can be released from the bacteria and bind to bacterial cell walls to mediate bacterial autolysis and eDNA release.
PrsA is a putative parvulin-type peptidyl-prolyl isomerase (PPIase) chaperone of S. mutans [Citation14]. PrsA homologs are present in diverse pathogens, including Listeria monocytogenes and Staphylococcus aureus, and are demonstrated to play important roles in the secretion and surface localization of virulence proteins to contribute to bacterial virulence and infection [Citation15–17]. Studies conducted on pathogenic streptococci also showed that PrsA regulates bacterial pathogenicity by modulating secretion of virulence factors [Citation18,Citation19]. In S. mutans, a prsA-mutant strain showed a marked increase in chain length, and reduced ability to produce the insoluble glucan and mutacin IV. Deletion of prsA also affects the surface localization of proteins, including glucosyltransferases and SpaP, and influenced the ability to form sucrose-dependent biofilms with aberrant architecture [Citation14,Citation20]. These findings provided information about the contribution of PrsA S. mutans virulence in the pathogenesis of dental caries, but whether PrsA mediates secretion or surface localization of bacterial virulence factors involved in IE pathogenesis is unclear.
In this study, we investigated the role of PrsA in IE pathogenesis in an animal model of endocarditis in rats. We showed that PrsA mediates secretion and surface localization of AtlA, which contributes to bacterial eDNA release and biofilm formation in S. mutans-induced endocarditis.
Materials and methods
Bacterial strains and plasmids
S.mutans GS5 wild-type, prsA-deficient (ΔprsA), and prsA-complemented (comΔprsA) strains were grown in BHI broth (Difco Laboratories Inc., Detroit, MI, USA). A shuttle plasmid containing the GFPuv sequence (pPDGFPuv) was used to generate green fluorescent protein (GFP)-tagged bacteria as previously described [Citation21]. Spectinomycin (500 μg/mL; pPDGFPuv), kanamycin (500 μg/mL; ΔprsA and comΔprsA), and chloramphenicol (5 μg/mL; comΔprsA) were used to select the colonies.
Construction of δprsa and comδprsa strains
To generate the deletion mutant strain of prsA, a ligation-PCR mutagenesis strategy with a promoterless kanamycin cassette was used as previously described [Citation22]. The upstream and downstream sequences of prsA and the promoterless kanamycin resistance gene fragment were amplified from S. mutans GS5 genomic DNA and the pALH124 plasmid, respectively, using the primers listed in [Citation23]. The primers were designed according to the S. mutans GS5 genome database (https://www.ncbi.nlm.nih.gov/genome/). PCR reaction products were digested with EcoRI and ligated with T4 ligase. The ligation was directly transformed into wild-type S. mutans GS5 or a clinical isolate, 4152, and the correct deletion mutant strain was selected by PCR amplification. The loss of prsA expression and that of genes downstream of prsA was confirmed by RT-PCR. For construction of the prsA-complemented strain (comΔprsA), the prsA promoter and prsA gene fragments were PCR-amplified using the primers listed in . The PCR products of the promoter and prsA gene were digested with XbaI and cloned into the pMC340B_Cm plasmid [Citation21]. The complementation plasmid was transformed into the ΔprsA strain to generate the prsA-complemented strain (comΔprsA). Complementation of prsA was screened by PCR amplification, and further confirmed by RT-PCR and western blotting.
Table 1. PCR primers used in the current study.
A rat model of S. mutans-induced IE
A modified rat model of experimental streptococcal endocarditis was used to investigate the IE pathogenesis as previously described [Citation24]. The conduct of animal experiments was approved by the Institutional Animal Care and Use Committee (National Taiwan University, Taipei, Taiwan). Briefly, an injury in the aortic valves was created by the insertion of a polyethylene tube with a stainless-steel wire embedded inside into the left carotid artery. Twenty-four hours later, rats were intravenously infected with 1 × 109 CFU of bacteria. At 24 h post-infection, the vegetation was harvested and weighed. To quantify bacterial colonization, the vegetation samples were homogenized by ultrasonication. The vegetation homogenates from six rats from each group were plated and the resulting colonies were counted. For the observation of biofilms by confocal laser scanning microscopy (CLSM; Leica TCS SP5), S. mutans GS5 wild-type and the mutant strains were transformed with pPDGFPuv [Citation21]. S. mutans (1 × 109 CFU) were intravenously infected into the tail veins of catheterized rats and the vegetation was harvested 24 h later. For each vegetation sample, three regions were evaluated by confocal microscopy, and a representative image is shown. To detect bacterial eDNA, the vegetation was stained with propidium iodide (PI) and the bacterial biofilms were observed by confocal microscopy (Leica TCS SP5).
Biofilm formation assays
The eDNA-dependent biofilm formation assay was performed as previously described [Citation7]. Briefly, the bacterial biofilms were grown by inoculating approximately 107 CFU of S. mutans in 200 μL defined M4 medium with or without the indicated concentration of S. mutans chromosome DNA or recombinant AtlA in a 96-well polystyrene microtiter plate. S. mutans chromosome DNA was extracted using a commercial kit (Geno Plus Genomic DNA Extraction kit; Viogene, Taipei, Taiwan), and the recombinant AtlA was purified using nickel chelating chromatography as previously described [Citation21]. For quantification, the biofilms were stained with 0.1% crystal violet dye, which was extracted using extraction buffer (20% methanol and 80% acetone). The absorbance of the extracted crystal violet dye (550 nM) was measured with a MicroELISA reader (Dynatech Corp., Alexandria, VA, USA). All the biofilm experiments were performed in triplicate and repeated at least three times. For the observation by CLSM, GS5 wild-type and mutant strains were transformed with pPDGFPuv and cultured in 24-well plates for 16 h at 37 °C. After gently washing three times with PBS, the biofilms that formed on the glass coverslip were fixed with 2% paraformaldehyde for 30 min and then stained with 10 μM PI for 10 min before observation with CLSM. Three different regions were evaluated for each biofilm sample and a representative region is shown.
Autolysis assay
Autolysis was performed as previously described. Bacterial cells in the exponential growth phase (OD550 = 0.9) were harvested by centrifugation. After washing three times with PBS, bacterial cells were resuspended in autolysis buffer [Citation25] with or without recombinant AtlA to OD550 = 0.9. Bacterial autolysis was monitored by measuring the OD550 of the cell suspension. All experiments were performed in triplicate and repeated at least three times.
Semiquantification of eDNA
Bacterial eDNA released into the culture medium was detected as previously described [Citation7]. Briefly, culture tubes of wild-type and isogenic mutant strains were agitated using a vortex mixer (Vortex Genie, Scientific Industries, Inc.) at 3,200 rpm for 30 sec to disrupt the biofilm, and the culture supernatants were harvested by centrifugation, followed by filtration using filters with 0.45 μm pore size to remove bacteria cells. The eDNA in the supernatant was semi-quantified by conventional PCR amplification (25 cycles) or quantified PCR (qPCR) analysis using specific bacterial 16S rRNA primers listed in . Culture medium containing S. mutans GS5 genomic DNA or without seeding bacteria was used as a positive and negative control, respectively. The experiments were performed in triplicate and repeated at least three times. For detecting bacterial eDNA inside the vegetation, a Gentra Puregene Tissue kit (Qiagen, Hilden, Germany) was used to extract the total DNA without lysing the bacteria [Citation7]. The extracted bacterial eDNA was then semi-quantified by PCR amplification (25 cycles) or qPCR analysis using specific bacterial 16S rRNA primers. Three rats were used for detection of bacterial eDNA, and each rat represented one independent experiment.
Immunofluorescence staining of AtlA exposure on bacterial surface
Briefly, the bacteria were fixed with 4% paraformaldehyde for 30 min and blocked with 3% bovine serum albumin in PBS for 1 hour. Samples were then incubated with a rabbit anti-AtlA antibody (1:200 dilution) for 2 hours [Citation21], followed by Texas red-conjugated anti-rabbit secondary antibodies (1:400 dilution) for 2 hours. AtlA exposure on bacterial surface was observed using Apotome 3D Super Resolution Microscopy (ZEISS, Oberkochen, Germany). Three different regions were evaluated for each sample and a representative region is shown.
Cell wall pulldown assay
Binding of AtlA in the culture medium to bacterial cell walls was detected by a cell wall pulldown assay that was performed as previously described with some modification [Citation13]. Briefly, the supernatant of overnight bacterial cultures was harvested by centrifugation and incubated with lyophilized cell walls on a rotator for 2 h. The lyophilized cell walls were prepared as previously described with modifications [Citation26]. Briefly, the exponential growth phase cells (OD550 = 0.9) were collected by centrifugation. The cell pellet was washed three times with iced PBS and resuspended in 5% SDS solution. After boiling for 30 minutes, the bacterial cells were broken with glass beads in a homogenizer (FastPrep FP120 homogenizer; Qbiogene Inc., Montreal, Canada). After the addition of DNase, RNase, trypsin, and protease K, the mixtures were incubated at 37°C for 2 hours. The cell wall fragments were washed with acetone and lyophilized. After the incubation with culture supernatant, the cell walls were collected by centrifugation, and washed three times with PBS buffer. After washing, the cell wall-binding proteins were dissolved in SDS sample buffer and analysed by western blotting using anti-AtlA antibodies [Citation21]. The experiment was performed three times and a representative result is shown.
Statistical analysis
For differences between more than two groups, a 1-way analysis of variance (ANOVA) with subsequent Bonferroni multiple-comparisons test was used, and a Kruskal–Wallis test followed by Dunn’s test was used for nonparametrically distributed data. Differences were considered to be statistically significant at P < 0.05.
Results
PrsA-Deficient mutant has reduced formation of vegetation and biofilm in vivo
PPIase chaperones, including PrsA, mediate secretion of bacterial virulence proteins to contribute to bacterial pathogenesis, including streptococci [Citation15]. Amino acid sequence alignment of PrsA from pathogenic streptococci shows good conservation among PrsA sequences (), suggesting that the role of PsrA is similar in streptococcal pathogenesis. To investigate the role PrsA in the pathogenesis of oral streptococci-induced IE, experimental IE rats were intravenously infected with wild-type or prsA-deficient (ΔprsA) mutant strains of S. mutans GS5, or a clinical blood isolate, 4152 or its isogenic prsA mutant. The vegetations on the heart valve were harvested 24 hr after infection. The PrsA-deficient mutant (ΔprsA) exhibited a significantly reduced ability to colonize the heart valve and form vegetation compared with the parental strain (, and Supplementary Fig. S1a-c). Complementation of the prsA gene into prsA-deficient strains (comΔprsA) restored the ability to form vegetation, thus confirming the role of PrsA (, and Supplementary Fig. S1a-c). To further investigate the role of PrsA in bacterial biofilm formation on heart valves in vivo, GFP-tagged wild-type, prsA-deficient mutant, and prsA-complemented strains were used. The prsA-deficient strain consistently showed reduced ability to form biofilms on heart valves compared with the wild-type and complementation strains ( and Supplementary Fig. S1d). These data suggested that PrsA plays a role in modulating the ability of bacteria to form biofilm and vegetation on heart valves in vivo.
Figure 1. PrsA amino acid sequence conservation among pathogenic streptococci. (a) Alignment of the PrsA amino acid sequences from pathogenic streptococci. The sequences were obtained from the NCBI database. A high degree of sequence conservation was observed. (b) Homology tree of PrsA amino acid sequences from pathogenic streptococci.
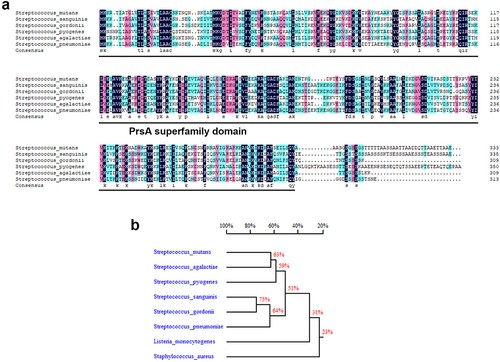
Figure 2. PrsA-Deficient mutant strain shows reduced ability to promote vegetation formation in a rat model of IE in rats. Investigation of the role of PrsA in the pathogenesis of IE using GS5 wild-type, ΔprsA, and comΔprsA strains in rat models of endocarditis. (a) Vegetation formation on the heart valves of endocarditis rats. Markers represent vegetation (black arrows) and valves (dashed lines). Scale bars represent 1 mm. (b and c) the number of colonized bacteria inside vegetation (b) and vegetation size (c) was measured. Data are presented as a scatter plot with mean ± standard error of the mean. **P < 0.001, *P < 0.05 by Kruskal–Wallis test with subsequent Dunn’s test; ns, not significant. (d) Three-dimensional structure of GFP-tagged S. mutans biofilms inside the vegetation that was harvested from injured heart valves and observed by confocal laser scanning microscopy (630× magnification). Bars indicate 50 μm.
PrsA mediates release of bacterial eDNA that contributes to bacterial biofilm formation in vitro and in vivo
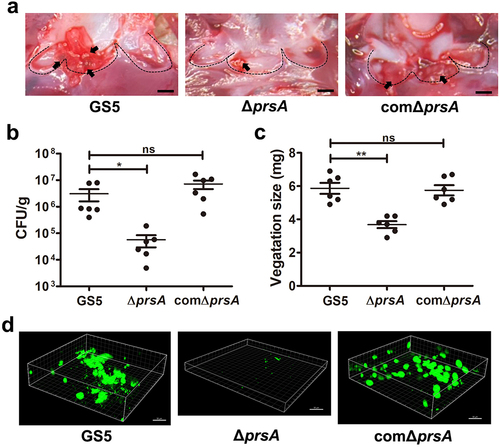
In vitro, the prsA-deficient mutant showed a similar growth rate to wild type, but had increased cell chain length ( and Supplementary Fig. S2a and S2b) that is similar to the atlA-deficient mutant strain [Citation25]. Our previous study indicated that S. mutans autolysin AtlA mediates autolysis and eDNA release to contribute to biofilm formation on the heart valves [Citation7]. Thus, we investigated the role of PrsA in eDNA-dependent biofilm formation. The prsA-deficient mutant showed reduced autolysis activity and reduced ability to release eDNA into the culture medium ( and Supplementary Fig. S2c and S2d). Moreover, the ability of the prsA-deficient mutant to release eDNA into the biofilm matrix and capacity for eDNA-dependent biofilm formation was diminished ( and Supplementary Fig. S2e-g). Addition of bacterial DNA to the culture medium of the prsA-deficient strain restored the bacterial biofilm formation capacity in a dose-dependent manner ( and Supplementary Fig. S2 h). These data confirmed the role of eDNA in PrsA-mediated biofilm formation.
Figure 3. PrsA mediates S. mutans cell separation, autolysis, eDNA release, and eDNA-dependent biofilm formation. (a) the bacterial growth was measured by detecting the absorbance of cultures at 550 nm. (b) Appearance of S. mutans GS5 wild-type, ΔprsA, and comΔprsA in the stationary phase were observed by light microscopy. (c) Bacterial autolysis assessed by measuring the OD550 of the cell suspensions. Data are expressed as the mean ± standard deviation of triplicate data. The values at 24 h were analyzed by 1-way ANOVA, ***P < 0.001. (d and e) Semi-quantitative analysis of bacterial eDNA release by conventional PCR and quantitative PCR (qPCR). Conventional PCR products obtained with 16S rRNA primers were analyzed on 1% agarose gels, and data of qPCR are represented as the mean of Ct values with SEM, and analyzed by 1-way ANOVA, *P < 0.001, *P < 0.05. Each spot represents the value from an independent culture. (f) Quantification of S. mutans biofilm formation using a crystal violet staining assay. Means of OD550 absorbance readings ± standard deviation of triplicate data are shown, and the data were statistically analyzed by 1-way ANOVA. **P < 0.01, ns, not significant. (g) Confocal laser scanning microscopy images of S. mutans biofilms (630× magnification). S. mutans GS5 wild-type and mutant strains were transformed with pPdgfpuv (green), and bacterial eDNA was stained with 10 μM propidium iodide (PI). GFP, green fluorescent protein. (h) Quantification of eDNA inside biofilms by detecting the fluorescence intensity of propidium iodide staining. The quantified values of the extended focus images of biofilms were determined using ImageJ software. The data were analyzed by 1-way ANOVA from three independent experiments and are presented as the mean value ± standard deviation. **P < 0.01, ***P < 0.001 by. ns, not significant. (i) Quantification of biofilm by crystal violet staining. S. mutans GS5 wild-type or ΔprsA were grown in culture medium with or without a concentration series of bacterial DNA. The data are presented as the mean ± standard deviation, and were analyzed by 1-way ANOVA. **P < 0.01, ***P < 0.001. The results for a representative experiment from three independent experiments are shown. The results of two other repeats were available in Supplemental Materials.
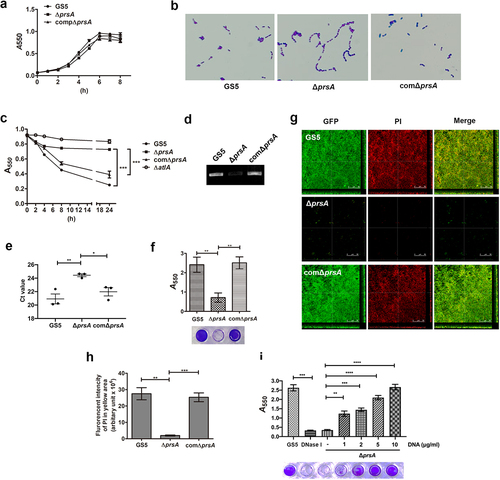
We next stained vegetation from samples from IE model rats with propidium iodide (PI) to observe bacterial eDNA release or bacterial lysis in situ by confocal microscopy ( and Supplementary Fig. S3a) [Citation7]. Colocalization of bacteria and DNA was observed (, and Supplementary Fig. S3a, yellow areas indicated by white arrows) for wild-type and prsA-complemented strains, suggesting that DNA was embedded inside the bacterial aggregates. Meanwhile, the prsA-deficient mutant had only small aggregates that lacked DNA (, and Supplementary Fig. S3a, red arrows). DNA inside the bacterial biofilm was further quantified by detecting the fluorescence intensity of DNA images in the yellow areas ( and Supplementary Fig. S3b). The DNA content inside biofilms formed by the prsA-deficient mutant was significantly reduced compared to wild-type and prsA-complemented strains ( and Supplementary Fig. S3b). To confirm reduction of eDNA inside the vegetation, we extracted the total DNA of the vegetations without lysing the bacteria [Citation7]. Semi-quantitative assays using 16S rRNA gene specific primers consistently showed that the bacterial eDNA levels in vegetation samples of the prsA-deficient mutant were significantly lower than that in samples of wild-type and the prsA-complemented strains (, Supplementary Fig. S3c and S3d) [Citation7]. These data suggested that PrsA mediates bacterial lysis or bacterial eDNA release that may contribute to biofilm formation in IE pathogenesis.
Figure 4. PrsA mediates in vivo eDNA-dependent biofilm formation. (a) Confocal laser scanning microscopic images of biofilm formation inside the vegetation (630× magnification). S. mutans GS5 wild-type and mutant strains were transformed with pPdgfpuv (green), and bacterial eDNA was stained with 10 μM propidium iodide (PI). GFP, green fluorescent protein. The red arrows indicate small bacterial aggregates without eDNA inside, and white arrows indicate the presence of both S. mutans and eDNA (yellow areas). The images shown are representative of three independent experiments. (b) Quantification of the eDNA inside the bacterial biofilms (yellow areas) by measuring the fluorescence intensity of propidium iodide staining. The quantified values of the extended focus images of biofilms were detected using ImageJ software. The data are presented as scatter plots with mean ± standard deviation. ***P < 0.001 by 1-way ANOVA. (c, d and e) Semi-quantification of bacterial eDNA inside the vegetation. Total DNA of vegetation was extracted without lysing bacteria, and the bacterial 16S rRNA gene was amplified by conventional PCR or quantitative PCR (qPCR) with specific primers. Conventional PCR products were analyzed on 1% agarose gels (c) and the intensity was semi-quantified using ImageJ software (d). Data of qPCR are represented as Ct values (e). These data were statistically analyzed by 1-way ANOVA and presented as scatter plots with means ± standard deviation from three independent experiments, ***P < 0.001, **P < 0.01, *P < 0.05.
PrsA contributes to the secretion and surface localization of AtlA
The characteristics of the prsA-deficient and atlA-deficient strains are similar in terms of enhanced cell chain length, reduced autolysis activity, reduced ability to release eDNA, and reduced biofilm forming capacity in vitro and in vivo [Citation7,Citation25]. Therefore, we investigated whether PrsA affects AtlA expression or function by extracting bacterial cell wall/membrane-associated proteins from total bacteria or biofilm populations with 4% SDS buffer and analysing the extracts by western blotting. The expression of cell wall/envelope-associated AtlA was similar for wild-type and mutant strains (, Supplementary Fig. S4a and S5a). However, in whole cell ELISA to detect cell surface exposure of bacterial proteins, AtlA exposure on the bacterial surface was reduced in the prsA-deficient mutant strain compared to the wild-type and prsA-complemented strains in both whole bacteria and biofilm populations (, Supplementary Fig. S4b and S5b). The result of immunofluorescence staining also showed that the reduced AtlA localization on bacterial surface of the prsA-deficient mutant strain (). In addition, the amount of secreted AtlA was dramatically reduced in the bacterial culture medium of the prsA-deficient mutant strain compared with the wild-type and prsA-complemented strains (, Supplementary Fig. S4c and S5c). A previous study indicated that AtlA binds to serotype c cell wall carbohydrates and mediates bacterial cell wall separation [Citation13]. To further confirm that secreted AtlA can bind to bacterial surfaces, we performed a cell wall pulldown assay with bacterial culture medium supernatants [Citation13] and used western blotting to examine the binding of secreted AtlA in the culture medium of wild-type or the prsA-complemented strain to bacterial cell walls (). Together, these data suggested that PrsA mediates secretion and surface localization of AtlA.
Figure 5. PrsA mediates surface exposure and secretion of AtlA. (a) Cell wall/envelope-associated proteins of S. mutans GS5 wild-type, ΔprsA, and comΔprsA were extracted with 4% sodium dodecyl sulfate (SDS) sample buffer. AtlA was detected by western blotting. (b and c) AtlA exposure on the bacterial surface detected by whole-cell ELISA (b) and immunofluorescence staining using anti-AtlA antibodies (c). Data of whole-cell ELISA are presented as the means ± standard deviation of triplicate data, and were analyzed by 1-way ANOVA. ***P < 0.001, ns, not significant. The result is a representative experiment from three independent experiments. The results of two other repeats were available in Supplemental Materials. (b). Immunofluorescence staining of AtlA on the bacterial surface was detected by ZEISS Apotome 3D Super Resolution Microscopy (c). Bar indicates 1μm. (d) AtlA secretion to the culture medium detected by western blotting. Cell-free overnight culture supernatants from S. mutans wild-type, ΔprsA, and comΔprsA were concentrated 20-fold, and analyzed by western blotting using anti-AtlA antibodies. (e) Cell wall pulldown of bacterial proteins in the culture supernatant of S. mutans wild-type, ΔprsA, and comΔprsA detected by western blotting using anti-AtlA antibodies.
Complementation of AtlA in the culture medium restored eDNA-dependent biofilm formation by the prsA-deficient mutant strain
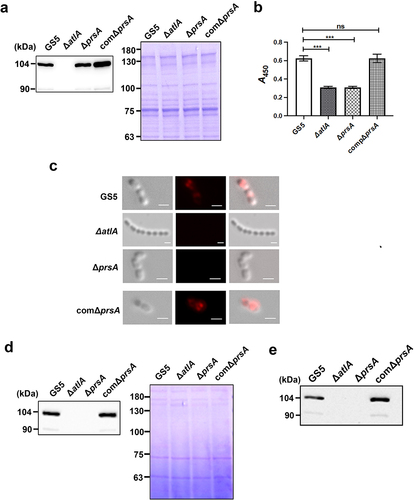
To further explore the role of secreted AtlA in PrsA-mediated biofilm formation, the recombinant AtlA (rAtlA) was used [Citation21]. Addition of rAtlA dose-dependently enhanced the autolysis activity of the prsA-deficient mutant strain (). Consistently, complementation with rAtlA in the culture medium dose-dependently restored eDNA release to the biofilm matrix as well as the capacity of the prsA-deficient mutant strain to form biofilms (. Treatment with DNase I reduced the rAtlA-enhanced biofilm formation of the prsA-deficient mutant strain, suggesting that addition of rAtlA in the culture bacterial medium indeed enhanced bacterial eDNA release that contributed to biofilm formation by the prsA-deficient mutant strain (). We next used the recombinant AtlA proteins having a C-terminal truncation (residues 167–776) or the putative AtlA C-terminal catalytic domain (residues 776–979) to understand whether AtlA enzyme activity was important for PrsA-mediated biofilm formation () [Citation7]. The autolysis activity of the recombinant full length and C-terminal domain of AtlA were confirmed using a zymogram assay (). Addition of recombinant full length or the C-terminal domain of AtlA restored the autolysis activity and eDNA-dependent biofilm formation of prsA-deficient mutant strain (). However, the C-terminal truncated mutant of AtlA had no effect. These data emphasized the important role of AtlA enzyme activity in PrsA-mediated biofilm formation and also confirmed the role of secreted AtlA in PrsA-mediated biofilm formation.
Figure 6. Addition of recombinant AtlA to the culture medium restores the capacity of the prsA-deficient mutant strain to form eDNA-dependent biofilm. (a) Bacterial autolysis assessed by measuring the OD550 of the cell suspension of the prsA-deficient mutant strain containing a concentration series of recombinant AtlA (rAtla). Data are expressed as the mean ± standard deviation of triplicate data; values at 24 h were analyzed by 1-way ANOVA, ***P < 0.001. (b) Quantification of biofilms of prsA-deficient mutant strain (ΔprsA) cultured in medium containing indicated concentrations of recombinant rAtla using a crystal violet staining assay. The experiment was performed in triplicate, and the data are presented as the mean ± standard deviation, and were analyzed by 1-way ANOVA. ***P < 0.001, ns, not significant. (c) Confocal laser scanning microscopy images of ΔprsA biofilms cultured in medium containing a series of rAtla concentrations (630× magnification). S. mutans GS5 wild-type and mutant strains were transformed with pPdgfpuv (green), and bacterial eDNA was stained with 10 μM propidium iodide (PI). GFP, green fluorescent protein; PI, propidium iodide. (d) Quantification of eDNA inside the biofilm by detecting the fluorescence intensity of propidium iodide staining. The quantified values of the extended focus images of biofilms were detected using ImageJ software, and statistically analyzed by 1-way ANOVA. The means ± standard deviation of three independent experiments is presented. ***P < 0.001, **P < 0.01, and *P < 0.05. (e) Quantification of S. mutans biofilm by a crystal violet assay. S. mutans GS5 wild-type and ΔprsA biofilms were grown in the culture medium with or without 20 μg/ml rAtla or 5 unit/ml DNase I. Means of OD550 absorbance readings ± standard deviation of triplicate data is shown; ***P < 0.001 by 1-way ANOVA. The results for a representative experiment from three independent experiments are shown. The results of two other repeats were available in Supplemental Materials.
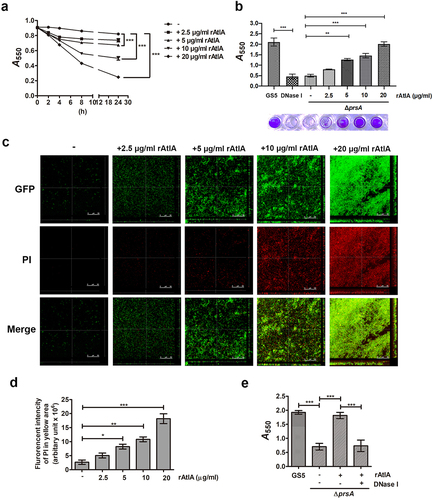
Figure 7. AtlA enzyme activity is important to PrsA-mediated bacterial autolysis and biofilm formation. (a) Schematic representation of the domain structure of AtlA. sp, signal peptide. (b) the virous recombinant AtlA proteins (rAtla) were separated by SDS-PAGE followed by silver staining. Lane1, full length of AtlA (residues 24–979); lane 2, mature form of AtlA (residues 167–979); 3, C-terminal truncation isoform (residues 167–776); lane 4, putative catalytic domain of AtlA (residues 776–979) (c) Zymographic analysis of rAtla enzyme activities. The rAtla proteins were separated by SDS-PAGE containing 1% (wet weight) heat-killed S. mutans cells. (d) Bacterial autolysis assessed by measuring the OD550 of the cell suspension of the prsA-deficient mutant strain containing 20 μg/ml indicated rAtla isoforms. The experiment was performed in triplicate and repeated three time. Data are expressed as the mean ± standard deviation of triplicate data. The values at 24 h were analyzed by 1-way ANOVA, ***P < 0.001. (e) Confocal laser scanning microscopy images of ΔprsA biofilms cultured in medium containing 20 μg/ml indicated rAtla isoforms (630× magnification). S. mutans GS5 wild-type and mutant strains were transformed with pPdgfpuv (green), and bacterial eDNA was stained with 10 μM propidium iodide (PI). GFP, green fluorescent protein; PI, propidium iodide. (f) Quantification of eDNA inside the biofilm by detecting the fluorescence intensity of propidium iodide staining. The quantified values of the extended focus images of biofilms were detected using ImageJ software, and statistically analyzed by 1-way ANOVA. The means ± standard deviation of three independent experiments is presented. ***P < 0.001. (g) Quantification of S. mutans biofilm by a crystal violet assay. S. mutans GS5 wild-type and ΔprsA biofilms were grown in the culture medium with or without 20 μg/ml indicated rAtla isoforms. Means of OD550 absorbance readings ± standard deviation of triplicate data are shown; ***P < 0.001, **P<0.01, by 1-way ANOVA. The results for a representative experiment from three independent experiments are shown. The results of two other repeats were available in Supplemental Materials.
Discussion
PrsA has a role in mediating virulence factor secretion and surface localization in many bacteria. Results of this study demonstrate for the first time that prsA mediates S. mutans virulence in a fatal disease, IE. Our previous studies demonstrated the important role of AtlA-mediated bacterial eDNA release in biofilm formation on damaged heart valves in IE [Citation7]. AtlA was also shown to bind to bacterial cell wall carbohydrates to mediate bacterial autolysis and cell division [Citation13]. Although a previous study indicated that PrsA also has a role in bacterial cell division, there was no direct evidence that indicated a relationship between PrsA and AtlA [Citation20]. In the present study, we provide evidence for an important role for PrsA in surface localization and release of AtlA that contributes to eDNA release and biofilm formation.
Bacterial PPIases are divided into three superfamilies: FKBPs, cyclophilins, and parvulins [Citation15]. PPIases such as PrsA modulate virulence in bacterial pathogens [Citation15]. PrsA is an N-terminally lipid-anchored lipoprotein that belongs to the parvulin family [Citation13,Citation27]. Located in the compartment between the cell membrane and cell wall, PrsA is thought to chaperone folding of bacterial virulence proteins destined for cell export [Citation13,Citation27]. Bacterial proteins exported to the plasma membrane by the Sec translocase are unfolded [Citation28]; thus, subsequent correct folding is critical for their secretion and function. PrsA located in the membrane–wall interface promotes appropriate protein folding by catalysing cis-trans isomerization of peptide bonds preceding prolyl residues [Citation15,Citation29]. In Bacillus subtilis, misfolded penicillin-binding proteins (PBP2a) were observed in the prsA mutant strain [Citation30], and in S. aureus PrsA functions not only in the secretion of proteases and phospholipase, but also affects the enzyme activity of these secreted virulence factors [Citation17]. Our data showed that in a ΔprsA-deficient mutant strain, AtlA retained its association with the bacteria cell wall/envelope, but had reduced autolysis activity (, Supplementary Fig. S2c and S5a). This data suggests an important role for PrsA in modulating the enzyme activity of AtlA on the bacterial cell wall. Our data also showed that PrsA mediates AtlA secretion (, Supplementary Fig. S4 and S5), and a recent study indicated that AtlA specifically binds to serotype c carbohydrates in the bacterial cell wall to mediate bacterial cell division and autolysis [Citation13]. Based on our findings and those that were previously described, we propose a hypothetical model in which PrsA located inside the cell wall regulates AtlA by chaperoning its folding and secretion (Supplementary Fig. S6). The secreted AtlA then sequentially binds to cell wall carbohydrates and mediates both cell autolysis and eDNA release that contributes to biofilm formation on damaged heart valves in IE.
Regarding S. mutans virulence, previous studies described several mutant strains that display similar phenotypes to the prsA-deficient mutant, including pcp-, liaR- or ropA-deficient mutant strains. These similarities include increased chain length and unchanged AtlA expression in the cell envelope/wall [Citation8,Citation20]. RopA is a bacterial PPIase that belongs to the FKBP family [Citation15], whereas LiaR and PCP are two regulatory proteins that are also involved in eDNA release and biofilm formation in IE pathogenesis [Citation8]. These earlier studies also showed that AtlA expression levels associated with the cell envelope were not changed in these isogenic mutant strains compared with the wild-type strain. Interestingly, our preliminary data indicated that AtlA secretion was reduced in the medium of pcp-, liaR- or ropA-deficient mutant strain cultures, but only the addition of rAtlA could restore the biofilm formation capacity of the ropA-deficient mutant strains (Supplementary Fig. S7). These data suggested that, similar to PrsA, S. mutans RopA may also play a role in the secretion of AtlA. RopA is a homolog of trigger factor, a ribosome-associated chaperone that is involved in the biogenesis and maturation of newly formed proteins [Citation31] and has been shown to contribute to the maturation and secretion of the S. pyogenes cysteine protease SspB [Citation32]. In S. mutans, inactivation of ropA reduced bacterial genetic competence as well as acid and peroxide tolerance, in addition to biofilm formation in the presence of saliva components [Citation33]. Our preliminary data showed a similar phenotype for the ropA- and prsA-deficient mutant strains. We will further investigate the actions of PrsA and RopA in the secretion of AtlA in future studies. We also observed a reduction in the amount of AtlA in the culture medium for liaR- and pcp- deficient mutant strains, but the addition of AtlA could not restore the phenotype, suggesting that, in addition to mediating AtlA secretion, LiaR and PCP may also influence AtlA function or affect bacterial autolysis machinery. Future studies should be performed to address how LiaR and PCP could affect AtlA function or bacterial autolysis machinery in IE pathogenesis.
In addition to IE, S. mutans is known to form biofilms on dental surfaces that are mediated by glucan synthesis catalysed by the glucosyltransferase (Gtf) system [Citation34,Citation35]. Previous studies also showed that eDNA crosslinks glucan to stabilize S. mutans biofilms on dental surfaces [Citation36] and that glucosyltransferase-I may also be involved in eDNA-dependent biofilm formation in vitro [Citation37]. Although reduced expression of glucosyltransferases and glucan production was also seen for the prsA-deficient mutans strain [Citation14], our data showed that addition of eDNA or rAtlA restored biofilm-forming capacity of the prsA-deficient S. mutans stain (), indicating an important role for eDNA and AtlA in prsA-mediated biofilm formation. In contrast to dental surfaces, there is no sucrose in plasma, and the glucosyltransferase-deficient strain GHS1DD, which showed reduced ability to form glucan-dependent biofilms relative to GS5 (Supplementary Fig. 8), had no reduction in the capacity to cause IE [Citation24]. This finding suggested that Gtfs and Gtf-mediated glucans may play limited roles in S. mutans (at least in GS5)-induced IE. However, we cannot exclude the role for glucan in other S. mutans strains or oral streptococci-induced IE. Indeed, previous studies indicated that bacterial binding to exopolysaccharides is important for immune evasion, as well as for the capacity to colonize heart valves in IE [Citation38,Citation39]. In addition, before entering circulation, oral streptococci may be enmeshed in sucrose-derived exopolysaccharides. Therefore, the role of PrsA-mediated glucan in the pathogenesis of IE requires further investigation.
The mechanism of biofilm formation on the heart valve in vivo is complex and involves bacterial immune evasion as well as adherence to endothelial cells and extracellular matrix proteins. Therefore, in addition to influencing AtlA secretion, PrsA may also affect the secretion or surface-localization of other bacterial virulence proteins that are involved in the pathogenesis of S. mutans-induced IE, and these putative virulence proteins require further investigation.
In conclusion, the results obtained in the present study demonstrated the important role of PrsA-mediated virulence in the pathogenesis of IE and that PrsA modulates the secretion and surface localization of AtlA to promote eDNA-dependent biofilm formation, which contributes, at least in part, to PrsA-mediated IE pathogenesis. Since autolysin-mediated eDNA-dependent biofilms are also observed in other species [Citation11,Citation40,Citation41], and several bacterial species carry PrsA homologues, the results obtained in this study could provide important information not only about the pathogenesis of S. mutans-induced IE, but could also enhance understanding of mechanisms associated with other infectious diseases that involve biofilm formation.
Supplemental Material
Download Zip (2.9 MB)Acknowledgments
This study was supported by the Ministry of Science and Technology (111-2320-B-038-063, 111-2320-B-002-066, 111-2320-B-002-076 and 108-2320-B-038-057-MY3).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2105351
Additional information
Funding
References
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108.
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39.
- Hoiby N, Bjarnsholt T, Givskov M, et al. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–332.
- Moreillon P, Que YA. Infective endocarditis. Lancet. 2004;363(9403):139–149.
- Jung CJ, Yeh CY, Shun CT, et al. Platelets enhance biofilm formation and resistance of endocarditis-inducing streptococci on the injured heart valve. J Infect Dis. 2012;205(7):1066–1075. DOI:10.1093/infdis/jis021
- Jung CJ, Yeh CY, Hsu RB, et al. Endocarditis pathogen promotes vegetation formation by inducing intravascular neutrophil extracellular traps through activated platelets. Circulation. 2015;131(6):571–581.
- Jung CJ, Hsu RB, Shun CT, et al. AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun. 2017;85(9). DOI:10.1128/IAI.00252-17
- Jung CJ, Hsu CC, Chen JW, et al. PspC domain-containing protein (PCP) determines Streptococcus mutans biofilm formation through bacterial extracellular DNA release and platelet adhesion in experimental endocarditis. PLoS Pathog. 2021;17(2):e1009289. DOI:10.1371/journal.ppat.1009289
- Ibanez de Aldecoa AL, Zafra O, Gonzalez-Pastor JE. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front Microbiol. 2017;8:1390.
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, et al. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487.
- Rice KC, Mann EE, Endres JL, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci, USA. 2007;104(19):8113–8118. DOI:10.1073/pnas.0610226104
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70(1):192–221.
- Zamakhaeva S, Chaton CT, Rush JS, et al. Modification of cell wall polysaccharide guides cell division in Streptococcus mutans. Nat Chem Biol. 2021;17(8):878–887. DOI:10.1038/s41589-021-00803-9
- Guo L, Wu T, Hu W, et al. Phenotypic characterization of the foldase homologue PrsA in Streptococcus mutans. Mol Oral Microbiol. 2013;28(2):154–165. DOI:10.1111/omi.12014
- Unal CM, Steinert M. Microbial Peptidyl-Prolyl cis / trans Isomerases (PPIases): Virulence factors and potential alternative drug targets. Microbiol Mol Biol Rev. 2014;78(3):544–571.
- Alonzo F 3rd, Freitag NE. Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect Immun. 2010;78(11):4944–4957.
- Wiemels RE, Cech SM, Meyer NM, et al. An intracellular peptidyl-prolyl cis / trans isomerase is required for folding and activity of the Staphylococcus aureus secreted virulence factor nuclease. J Bacteriol. 2017;199(1). DOI:10.1128/JB.00453-16
- Olsen RJ, Sitkiewicz I, Ayeras AA, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S a. 2010;107(2):888–893. DOI:10.1073/pnas.0911811107
- Liu H, Fu H, Jiang X, et al. PrsA contributes to Streptococcus suis serotype 2 pathogenicity by modulating secretion of selected virulence factors. Vet Microbiol. 2019;236:108375.
- Crowley PJ, Brady LJ. Evaluation of the effects of Streptococcus mutans chaperones and protein secretion machinery components on cell surface protein biogenesis, competence, and mutacin production. Mol Oral Microbiol. 2016;31(1):59–77.
- Jung CJ, Zheng QH, Shieh YH, et al. Streptococcus mutans autolysin AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol Microbiol. 2009;74(4):888–902.
- Lau PC, Sung CK, Lee JH, et al. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49(2):193–205.
- Kremer BH, van der Kraan M, Crowley PJ, et al. Characterization of the sat operon in Streptococcus mutans : Evidence for a role of ffh in acid tolerance. J Bacteriol. 2001;183(8):2543–2552.
- Shun CT, Lu SY, Yeh CY, et al. Glucosyltransferases of viridans streptococci are modulins of interleukin-6 induction in infective endocarditis. Infect Immun. 2005;73(6):3261–3270.
- Shibata Y, Kawada M, Nakano Y, et al. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect Immun. 2005;73(6):3512–3520.
- Severin A, Horne D, Tomasz A. Autolysis and cell wall degradation in a choline-independent strain of Streptococcus pneumoniae. Microb Drug Resist. 1997;3(4):391–400.
- Gothel SF, Marahiel MA. Peptidyl-Prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55(3):423–436.
- Sarvas M, Harwood CR, Bron S, et al. Post-Translocational folding of secretory proteins in gram-positive bacteria. Biochim Biophys Acta. 2004;1694(1–3):311–327.
- Fanghanel J, Fischer G. Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front Biosci. 2004;9(1–3):3453–3478.
- Hyyrylainen HL, Marciniak BC, Dahncke K, et al. Penicillin-Binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in bacillus subtilis. Mol Microbiol. 2010;77(1):108–127. DOI:10.1111/j.1365-2958.2010.07188.x
- Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone trigger factor. Biochim Biophys Acta. 2010;1803(6):650–661.
- Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. Embo J. 1998;17(21):6263–6275.
- Wen ZT, Suntharaligham P, Cvitkovitch DG, et al. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun. 2005;73(1):219–225.
- Mitchell TJ. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol. 2003;1(3):219–230.
- Bowen WH, Koo H. Biology of Streptococcus mutans- derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69–86.
- Rainey K, Michalek SM, Wen ZT, et al. Glycosyltransferase-Mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl Environ Microbiol. 2019;85(5). DOI:10.1128/AEM.02247-18
- Nagasawa R, Sato T, Nomura N, et al. Potential risk of spreading resistance genes within extracellular-DNA-dependent biofilms of Streptococcus mutans in response to cell envelope stress induced by sub-MICs of bacitracin. Appl Environ Microbiol. 2020;86(16). DOI:10.1128/AEM.00770-20
- Pulliam L, Dall L, Inokuchi S, et al. Effects of exopolysaccharide production by viridans streptococci on penicillin therapy of experimental endocarditis. J Infect Dis. 1985;151(1):153–156.
- Nguyen HTT, Nguyen TH, Otto M. The staphylococcal exopolysaccharide PIA - biosynthesis and role in biofilm formation, colonization, and infection. Comput Struct Biotechnol J. 2020;18:3324–3334.
- Qin Z, Ou Y, Yang L, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology (Reading). 2007;153:2083–2092.
- Liu Y, Burne RA. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol. 2011;193(11):2826–2837.
