ABSTRACT
Guanylate binding proteins (GBPs) are interferon-inducible cellular factors that belong to the superfamily of guanosine triphosphatases (GTPases) and play important roles in the cell-intrinsic defense against bacteria, protozoa and viruses. In a recent report in Cell Host & Microbe, we identify GBP5 as novel restriction factor of HIV-1 that reduces the infectivity of progeny virions by interfering with processing and incorporation of the viral envelope (Env) glycoprotein. The inhibitory activity of GBP5 requires C-terminal isoprenylation, mediating Golgi-association, but not its GTPase function. Notably, GBP5 expression levels vary considerably in human macrophages and inversely correlate with infectious virus yield. We demonstrate that GBP5 can be evaded by an unusual tradeoff mechanism: Naturally occurring mutations in the start codon of the viral accessory gene vpu attenuate GBP5 inhibition by increasing Env expression at the cost of Vpu function. Whether direct counteraction mechanisms or more subtle changes balancing Vpu and Env expression also affect HIV-1 inhibition by GBP5 remains to be clarified. Other open questions are whether GBP5 restricts HIV-1 in CD4+ T cells and if other GBP family members also decrease infectivity of HIV and/or additional enveloped viruses.
Restriction factors are host-encoded proteins that inhibit viral pathogens by targeting various steps in the viral replication cycle.Citation1,2 They represent key effectors of antiviral immune responses and play an important role as barriers to cross-species transmissions and in limiting viral spread.Citation3,4 Despite their enormous structural and functional diversity, restriction factors frequently share several characteristics: Most of them are cell-intrinsic proteins whose expression is induced or further upregulated by interferons (IFNs) in response to invading pathogens.Citation5 Furthermore, restriction factors frequently interact with viral proteins because they target the viral replication cycle and/or because they are counteracted by viral antagonists.Citation4,6 As a result, restriction factors are in a constant arms race with viruses and typically display signatures of strong positive selection pressure.Citation7 We recently used these characteristics as distinctive features in a genome-wide screen to identify previously uncharacterized antiretroviral proteins.Citation8 One interesting candidate identified was guanylate binding protein 5 (GBP5). This factor belongs to the superfamily of IFN-inducible guanosine triphosphatases (GTPases) comprising 4 subfamilies: guanylate binding proteins (GBPs), immunity-related GTPases (IRGs), very large inducible GTPases (VLIGs) and myxovirus resistance (Mx) proteins.Citation9 Members of these protein families are well-known to participate in cell-intrinsic immunity against bacteria, protozoa and viruses.Citation9 The human MxA protein, for example, is a well-established restriction factor of influenza A viruses.Citation10 Additionally, growing evidence highlights the role of IFN-inducible GTPases in the control of HIV-1 infection. For example, the interferon-inducible myxovirus resistance protein B (MxB) inhibits HIV-1 replication by targeting the viral capsid, resulting in impaired integration of viral DNA into the host genome.Citation11-13 However, the relative contribution of MxB to IFN-induced HIV restriction is still controversial.Citation14 In contrast to MxB, GBP5 targets the viral envelope (Env) glycoprotein to reduce infectivity of progeny virions.Citation8,15
Reduced virion infectivity in the presence of GBP5 correlates with impaired Env processing and incorporation.Citation15 The Env precursor gp160 is produced on the rough endoplasmic reticulum, where N-linked oligosaccharide side chains are attached cotranslationally.Citation16 During anterograde trafficking through the Golgi apparatus to the cell membrane, gp160 trimerizes and is cleaved by cellular proteases into the surface glycoprotein gp120 and the transmembrane glycoprotein gp41.Citation17 Mature gp120/gp41 trimers assemble at the plasma membrane and are incorporated into budding virions, where they mediate infection of new target cells. GBP5 decreases the levels of mature gp120 while increasing incorporation of unprocessed gp160 into progeny virions and thus impairs their ability to fuse with target cells. Our findings suggest that GBP5 targets Env processing in the Golgi apparatus and thereby impairs trafficking to the cell surface and subsequent incorporation.Citation15 GBP5 may exert broad antiretroviral activity as it also reduces infection mediated by the envelope glycoprotein of murine leukemia virus (MLV).Citation15 However, vesicular stomatitis virus glycoprotein (VSV-G)-dependent infection is not affected by GBP5.Citation15
Notably, 2 other cellular factors, 90K and membrane-associated-RING-CH8 (MARCH8), have also been shown to impair HIV-1 infectivity by targeting the Env glycoprotein ().Citation18,19 90K is an interferon-stimulated factor belonging to the family of scavenger receptor cysteine-rich (SRCR) domain-containing proteins that is upregulated in individuals with cancer or HIV-1 infection.Citation20 Cell-associated 90K, but not its secreted form, reduces HIV-1 infectivity by decreasing proteolytic processing of Env and virion-association of mature gp120 and gp41.Citation18 Similarly, MARCH8 decreases surface expression and incorporation of retroviral Envs, but additionally also targets VSV-G.Citation19 MARCH8 is a RING-finger E3 ubiquitin ligase that downregulates various transmembrane proteins from the cell surface. It has been suggested that recognition of entire 3-dimensional structures of transmembrane domains, rather than specific motifs, might confer broad antiviral activity to MARCH8.Citation19 The interplay between GBP5, 90K and MARCH8 in inhibiting HIV-1 and potentially other enveloped viruses, as well as their relative contribution to antiretroviral immunity require further investigation.
Figure 1. Antiviral factors targeting the Env glycoprotein. GBP5 and 90K reduce the infectivity of progeny virions by impaired processing of the gp160 precursor, resulting in reduced incorporation of mature gp120/gp41.Citation15,18 The membrane-associated protein MARCH8 downmodulates Env from the cell surface, thereby preventing its incorporation into budding virions.Citation19
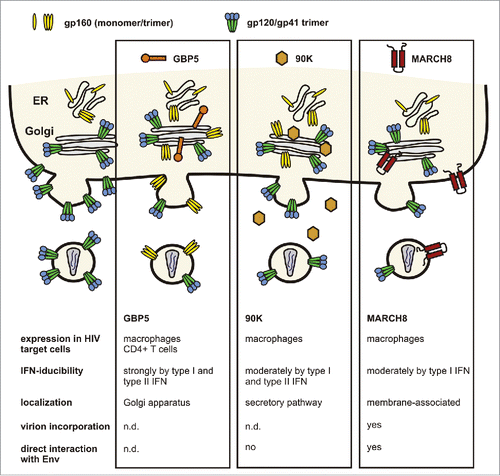
Expression of host restriction factors is commonly induced as part of the IFN-response to invading viruses.Citation21 90K and MARCH8 are predominantly expressed in macrophages and only slightly inducible by IFNs.Citation18,19 In contrast, GBP5 is expressed and strongly inducible by type I and II IFNs in macrophages as well as in CD4+ T cells, the main target cells of HIV. Notably, also HIV-1 infection induces GBP5 expression. This is in agreement with a correlation between GBP5 transcripts and viral RNA loads in infected individuals.Citation15 We noted that basal GBP5 expression levels vary considerably in macrophages from different blood donors. Strikingly, infectious virus yield inversely correlates with GBP5 expression levels over several orders of magnitude. siRNA-mediated knockdown confirmed that GBP5 inhibits HIV-1 replication in these cells by reducing the infectivity of progeny viruses.Citation15 It is long known that the susceptibility of macrophages to HIV-1 replication varies substantially between infected individuals. This phenomenon could only in part be attributed to different levels of the viral CD4 receptor and the presence of a 32 base pair deletion in the ccr5 gene that reduces CCR5 coreceptor expression.Citation22-24 Our findings provide a plausible explanation for the large donor variation in the susceptibility of macrophages to HIV-1 infection.Citation15
In humans, 7 closely related paralogs have been identified within the family of guanylate binding proteins (GBP1–7).Citation25 These 67–73 kDa proteins exhibit a bidomain architecture comprising a globular N-terminal G-domain and a C-terminal helical domain (CTHD).Citation9 The G-domain harbors the GTPase activity, which allows hydrolysis of GTP to GDP and GMP.Citation26,27 Additionally, GBP1, GBP2 and GBP5 contain an isoprenylation motif (CaaX) at their C-terminus, where lipid moieties can be attached to anchor these cytosolic proteins to membranes of the Golgi apparatus.Citation27,28 Our mutational analyses show that the GTPase function of GBP5 is dispensable for its ability to inhibit HIV-1. However, an isoform of GBP5 lacking 97 amino acids at its C-terminus, as well as a mutant containing a cysteine to alanine substitution at amino acid position 583 that prevents isoprenylation, lack antiviral activity.Citation15 This is conceivable as isoprenylation allows localization of GBP5 in membranes of the Golgi apparatus, the site of Env processing.
The human gbp cluster originated from gene duplication events and all human GBPs exhibit high sequence homology.Citation25 It will therefore be interesting to investigate whether other members of the human GBP family share the antiretroviral properties of GBP5. GPB1, the closest relative of GBP5, contains a C-terminal isoprenylation motif and has been reported to exert antiviral activity against dengue, hepatitis C, vesicular stomatitis and encephalomyocarditis viruses.Citation29-31 However, GBP1 does not inhibit HIV-1.Citation15 GBP2 is another potential antiretroviral effector because it shares a C-terminal CxxL motif with GBP5 that serves as attachment signal for geranylgeranyl moieties. In contrast, the C-terminal CaaS motif in GBP1 is farnesylated.Citation28 Since the identity of the lipid moiety co-determines the subcellular localization of GBPs,Citation27 geranylgeranylation might be critical for antiretroviral activity. It will also be interesting to analyze the evolutionary conservation of GBP5-mediated restriction. In silico analyses of the gbp locus revealed that several gene duplication and gene loss events occurred during mammalian evolution. As a result, various species differ in the presence of distinct gbp paralogs within this gene cluster. Thus, it is of interest to clarify whether gain of antiviral activity by other GBP family members might have removed the selective pressure to maintain gbp5 in species that lack expression of this paralog, and whether viruses derived from these species are particularly susceptible to this restriction factor.
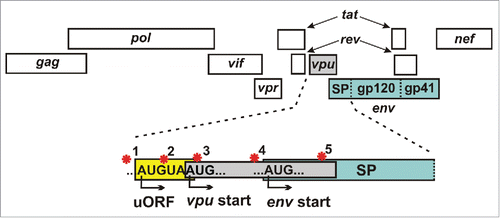
One common feature of relevant restriction factors is that viruses evade them by evolving escape mutations or direct counteraction strategies.Citation4,6 To achieve the latter, primate lentiviruses express a variety of so-called accessory proteins (Vif, Vpr, Vpu, Nef and Vpx).Citation4 Some of the respective genes partially overlap with other open reading frames in the viral genome. For example, the 3′-end of HIV-1 vpu overlaps with env and both proteins are expressed from a bicistronic mRNA (). Thus, Env expression depends on leaky ribosomal scanning and/or ribosomal shunting.Citation32,33 Accordingly, previous studies have shown that naturally occurring mutations in the vpu start codon increase Env expression by allowing immediate translation of env.Citation34 We confirmed this and further showed that increased expression and incorporation of Env into viral particles reduces their sensitivity to GBP5 inhibition.Citation15 Thus, GBP5 can be counteracted by an unusual tradeoff mechanism, in which Env expression is increased at the cost of Vpu functions.Citation15
Figure 2. Regulation of env expression. Simplified depiction of the HIV-1 genome highlighting different regulatory mechanisms (red stars) to modulate env expression from bicistronic vpu/env mRNAs: (1) The non-coding rev1-vpu intergenic region determines the strength of the vpu Kozak sequence and balances vpu and env expression.Citation43,44 (2) The minimal open reading frame (ORF) upstream of the vpu start codon contributes to efficient downstream env translation.Citation43 (3) A defective vpu start codon allows immediate translation of env.Citation34 (4) The Kozak consensus upstream of the env start codon determines transcription efficiency. (5) A positively charged amino acid at position 12 of the Env signal peptide (SP) is associated with efficient Env production and trafficking in transmitted-founder viruses.Citation45-47
Mutations in the vpu initiation codon increasing Env expression are usually associated with significant disadvantages for the virus as Vpu counteracts the restriction factor tetherin and degrades CD4 to enable efficient release of budding virions.Citation2 Furthermore, Vpu inhibits activation of the transcription factor NF-κB to suppress expression of antiviral genes.Citation35 Finally, increased expression and virion incorporation of Env might hamper viral replication due to increased susceptibility to neutralizing antibodies and antibody-dependent cell-mediated cytotoxicity (ADCC).Citation36 Nevertheless, mutations in the vpu initiation codon are found at an unusual high frequency, particularly in macrophage-tropic HIV-1 strains.Citation34,37,38,39 Thus, vpu-mutations that reduce viral susceptibility to GBP5 might be beneficial in specific cell types and body compartments associated with a reduced selective pressure for Vpu function and low Env expression. Vpu-mediated degradation of CD4, for example, might be less critical in macrophages, since these cells only express low levels of this receptor.Citation40 Furthermore, lack of Vpu-mediated tetherin antagonism increases the number of virions that are retained at the cell surface and may thereby facilitate direct cell-to-cell transmission of HIV-1.Citation41 In immunosecluded environments, such as the brain, lack of Vpu-mediated NFκ-B inhibition might even be beneficial for the virus. NF-κB is not only important for antiviral gene expression but also a key enhancer of HIV-1 gene expression via the long-terminal repeat (LTR) promoter.Citation42 Lack of Vpu-mediated NF-κB inhibition might thus increase LTR activity and virus production.Citation35 This might explain why vpu-deficient HIV-1 strains replicate efficiently in macrophages and produce similar amounts of cell-free virus as their counterparts with intact vpu genes despite reduced virion release efficiency.Citation15,34,38
Complete loss of Vpu expression is only the extreme of many alterations that might affect the balance between vpu and env expression and hence the susceptibility of HIV-1 to GBP5 inhibition. For example, a minimal upstream open reading frame (uORF) consisting only of a start and stop codon that partially overlaps with the vpu transcription start site has been shown to enhance ribosomal access to the env start codon ().Citation43 Furthermore, changes in the strength of the vpu upstream noncoding nucleotides (Kozak consensus), as well as mutations in the rev1-vpu intergenic region might influence env expression levels.Citation43,44 In addition, it has been shown that transmitted-founder (TF) virions are more infectious and contain about twice as much Env as chronic HIV variants.Citation45 These differences were ascribed to specific sequence signatures in the signal peptide of Env. While a positively charged amino acid at position 12 in TF strains was associated with high Env expression and trafficking, non-basic residues were enriched in viruses from chronically infected individuals.Citation46,47 It will be interesting to determine possible correlations between alterations in the HIV-1 genome enhancing Env expression and the levels of GBP5, 90K and MARCH8 expression in HIV-1 infected individuals.
While it has been established that HIV-1 particles contain only approximately 10 Env spikes,Citation16 surprisingly little is known about the levels of Env expression and virion incorporation by other primate lentiviruses. Notably, a vpu gene is only found in HIV-1 and its SIV precursors from chimpanzees and gorillas, as well as closely related SIVs infecting greater-spot nosed monkeys, mustached monkeys, mona and dent's mona monkeys. Thus, most primate lentiviruses do not need to balance Vpu and Env expression and it remains to be determined whether primate lentiviruses that do not encode vpu are less sensitive to GBP5 because they might express higher levels of Env.
Several highly relevant restriction factors, such as APOBEC3G, Tetherin, SAMHD1 and SERINC5, are directly antagonized by the accessory proteins Vif, Vpu, Vpx and Nef of primate lentiviruses.Citation2 Whether GBP5 is also directly counteracted requires further investigation. It has long been known that the accessory protein Vpr promotes HIV-1 replication in macrophages,Citation48 and more recently, it has been suggested that Vpr overcomes a macrophage-specific restriction of Env expression and virion production.Citation49 However, it is unclear if Vpr directly antagonizes an antiviral effector or rather inhibits the induction of a restriction factor that targets Env and is induced as part of the innate antiviral IFN response.Citation49,50 Although it is unlikely that HIV-1 Vpr counteracts GBP5, since it efficiently inhibits virion infectivity in the presence of Vpr,Citation15 these findings add to the accumulating evidence that retroviral Env is a suitable target for antiviral proteins.Citation15,18,19 Thus far, however, HIV-1 inhibition by targeting Env glycoproteins has only been demonstrated in macrophages.Citation15,18,19,49 Since GBP5 is also efficiently expressed and further inducible in CD4+ T cells,Citation15 it will be interesting to determine whether it also restricts retroviral infectivity in this major HIV-1 target cell type.
The identification of GBP5 as a relevant effector of the antiretroviral IFN-response adds to the growing evidence that IFN-inducible guanosine triphosphatases play an important role in innate immune responses against various pathogens.Citation9-13,15 Furthermore, it is striking that 2 GTPases, i.e. MxB and GBP5, inhibit HIV-1 replication by targeting different steps of the viral replication cycle and that GTPase activity seems dispensable in both cases.Citation11,13,15 Thus, further studies to fully define the evolution, relevance and antiviral mechanisms of IFN-inducible GTPases are clearly warranted.
Abbreviations
| ADCC | = | Antibody-dependent cell-mediated cytotoxicity |
| CTHD | = | C-terminal helical domain |
| GBP | = | Guanylate binding protein |
| GDP | = | Guanosine diphosphate |
| GMP | = | Guanosine monophosphate |
| GTP | = | Guanosine triphosphate |
| GTPase | = | Guanosine triphosphatase |
| gp | = | Glycoprotein |
| HIV | = | Human immunodeficiency virus |
| IFN | = | Interferon |
| IRG | = | Immunity-related GTPase |
| LTR | = | Long terminal repeat |
| MARCH8 | = | Membrane-associated-RING-CH8 |
| MLV | = | Murine leukemia virus |
| MxB | = | Myxovirus resistance protein B |
| Nef | = | Negative factor |
| Rev | = | Regulator of virion |
| SIV | = | Simian immunodeficiency virus |
| SRCR | = | Scavenger receptor cysteine-rich |
| TF | = | Transmitted-founder |
| uORF | = | Upstream open reading frame |
| Vif | = | Viral infectivity factor |
| VLIG | = | Very large inducible GTPase |
| Vpr | = | Viral protein r |
| Vpu | = | Viral protein unknown/Viral protein unique to HIV-1 |
| Vpx | = | Viral protein x |
| VSV-G | = | Vesicular stomatitis virus glycoprotein |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dré van der Merwe for critical proof-reading of the manuscript.
Funding
DH is funded by the International Graduate School in Molecular Medicine, Ulm. FK is supported by grants from the Deutsche Forschungsgemeinschaft (DFG), European FP7 “HIT HIDDEN HIV” (305762), and an Advanced ERC Investigator Grant.
References
- Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who's interfering with whom? Nat Rev Micro 2015; 13:403-13; http://dx.doi.org/10.1038/nrmicro3449
- Kluge SF, Sauter D, Kirchhoff F. SnapShot: antiviral restriction factors. Cell 2015; 163:774-4.e1; PMID:26496613; http://dx.doi.org/10.1016/j.cell.2015.10.019
- Krupp A, McCarthy KR, Ooms M, Letko M, Morgan JS, Simon V, Johnson WE. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLOS Pathog 2013; 9:e1003641; PMID:24098115; http://dx.doi.org/10.1371/journal.ppat.1003641
- Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 2010; 8:55-67; PMID:20638642; http://dx.doi.org/10.1016/j.chom.2010.06.004
- Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2012; 2:a006940; PMID:22553496; http://dx.doi.org/10.1101/cshperspect.a006940
- Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem 2012; 287:40875-83; PMID:23043100; http://dx.doi.org/10.1074/jbc.R112.416925
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 2012; 46:677-700; PMID:23145935; http://dx.doi.org/10.1146/annurev-genet-110711-155522
- McLaren PJ, Gawanbacht A, Pyndiah N, Krapp C, Hotter D, Kluge SF, Götz N, Heilmann J, Mack K, Sauter D, et al. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 2015; 12:41; PMID:25980612; http://dx.doi.org/10.1186/s12977-015-0165-5
- Kim B-H, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in Host Defense. Cell Host Microbe 2012; 12:432-44; PMID:23084913; http://dx.doi.org/10.1016/j.chom.2012.09.007
- Haller O. Dynamins Are Forever: MxB Inhibits HIV-1. Cell Host Microbe 2013; 14:371-3; PMID:24139395; http://dx.doi.org/10.1016/j.chom.2013.10.002
- Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 2013; 502:563-6; PMID:24121441; http://dx.doi.org/10.1038/nature12653
- Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 2013; 14:398-410; PMID:24055605; http://dx.doi.org/10.1016/j.chom.2013.08.015
- Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, Hué S, Barclay WS, Schulz R, Malim MH. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 2013; 502:559-62; PMID:24048477; http://dx.doi.org/10.1038/nature12542
- Opp S, Vieira DASA, Schulte B, Chanda SK, Diaz-Griffero F. MxB Is Not Responsible for the Blocking of HIV-1 Infection Observed in Alpha Interferon-Treated Cells. J Virol 2015; 90:3056-64; PMID:26719253; http://dx.doi.org/10.1128/JVI.03146-15
- Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Stürzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, et al. Guanylate Binding Protein (GBP) 5 Is an Interferon-Inducible Inhibitor of HIV-1 Infectivity. Cell Host Microbe 2016; 19:504-14.
- Checkley MA, Luttge BG, Freed EO. HIV-1 Envelope Glycoprotein Biosynthesis, Trafficking, and Incorporation. J Mol Biol 2011; 410:582-608; PMID:21762802; http://dx.doi.org/10.1016/j.jmb.2011.04.042
- Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 1992; 360:358-61; PMID:1360148; http://dx.doi.org/10.1038/360358a0
- Lodermeyer V, Suhr K, Schrott N, Kolbe C, Stürzel CM, Krnavek D, Münch J, Dietz C, Waldmann T, Kirchhoff F, et al. 90K, an interferon-stimulated gene product, reduces the infectivity of HIV-1. Retrovirology 2013; 10:111; PMID:24156545; http://dx.doi.org/10.1186/1742-4690-10-111
- Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, Fujita H, Tokunaga K. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat Med 2015; 21:1502-7; PMID:26523972; http://dx.doi.org/10.1038/nm.3956
- Goffinet C. Cellular Antiviral Factors that Target Particle Infectivity of HIV-1. Curr HIV Res 2016; 14:211-6; PMID:26674651; http://dx.doi.org/10.2174/1570162X14666151216145521
- Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, Shianna KV, Ge D, Günthard HF, Goldstein DB, et al. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog 2010; 6:e1000781; PMID:20195503; http://dx.doi.org/10.1371/journal.ppat.1000781
- Bol SM, van Remmerden Y, Sietzema JG, Kootstra NA, Schuitemaker H, van 't Wout AB. Donor variation in in vitro HIV-1 susceptibility of monocyte-derived macrophages. Virology 2009; 390:205-11; PMID:19535121; http://dx.doi.org/10.1016/j.virol.2009.05.027
- Naif HM, Li S, Alali M, Chang J, Mayne C, Sullivan J, Cunningham AL. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocyte-derived macrophages by using twins. J Virol 1999; 73:4866-81; PMID:10233948
- Pesenti E, Pastore C, Lillo F, Siccardi AG, Vercelli D, Lopalco L. Role of CD4 and CCR5 levels in the susceptibility of primary macrophages to infection by CCR5-dependent HIV type 1 isolates. AIDS Res Hum Retroviruses 1999; 15:983-7; PMID:10445810; http://dx.doi.org/10.1089/088922299310494
- Olszewski MA, Gray J, Vestal DJ. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J Interferon Cytokine Res 2006; 26:328-52; PMID:16689661; http://dx.doi.org/10.1089/jir.2006.26.328
- Schwemmle M, Staeheli P. The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J Biol Chem 1994; 269:11299-305; PMID:7512561
- Tripal P, Bauer M, Naschberger E, Mörtinger T, Hohenadl C, Cornali E, Thurau M, Stürzl M. Unique features of different members of the human guanylate-binding protein family. J Interferon Cytokine Res 2007; 27:44-52; PMID:17266443; http://dx.doi.org/10.1089/jir.2007.0086
- Modiano N, Lu YE, Cresswell P. Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-gamma-inducible cofactor. Proc Natl Acad Sci USA 2005; 102:8680-5; PMID:15937107; http://dx.doi.org/10.1073/pnas.0503227102
- Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 1999; 256:8-14; PMID:10087221; http://dx.doi.org/10.1006/viro.1999.9614
- Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, et al. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 2009; 50:1727-37; PMID:19821486; http://dx.doi.org/10.1002/hep.23195
- Pan W, Zuo X, Feng T, Shi X, Dai J. Guanylate-binding protein 1 participates in cellular antiviral response to dengue virus. Virol J 2012; 9:292; PMID:23186538; http://dx.doi.org/10.1186/1743-422X-9-292
- Guerrero S, Batisse J, Libre C, Bernacchi S, Marquet R, Paillart J-C. HIV-1 replication and the cellular eukaryotic translation apparatus. Viruses 2015; 7:199-218; PMID:25606970; http://dx.doi.org/10.3390/v7010199
- Anderson JL, Johnson AT, Howard JL, Purcell DFJ. Both Linear and Discontinuous Ribosome Scanning Are Used for Translation Initiation from Bicistronic Human Immunodeficiency Virus Type 1 env mRNAs. J Virol 2007; 81:4664-76; PMID:17329338; http://dx.doi.org/10.1128/JVI.01028-06
- Schubert U, Bour S, Willey RL, Strebel K. Regulation of virus release by the macrophage-tropic human immunodeficiency virus type 1 AD8 isolate is redundant and can be controlled by either Vpu or Env. J Virol 1999; 73:887-96; PMID:9882289
- Sauter D, Hotter D, Van Driessche B, Stürzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier J-C, et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep 2015; 10:586-99; PMID:25620704; http://dx.doi.org/10.1016/j.celrep.2014.12.047
- von Bredow B, Arias JF, Heyer LN, Gardner MR, Farzan M, Rakasz EG, Evans DT. Envelope Glycoprotein Internalization Protects Human and Simian Immunodeficiency Virus-Infected Cells from Antibody-Dependent Cell-Mediated Cytotoxicity. J Virol 2015; 89:10648-55; PMID:26269175; http://dx.doi.org/10.1128/JVI.01911-15
- Thomas ER, Dunfee RL, Stanton J, Bogdan D, Kunstman K, Wolinsky SM, Gabuzda D. High frequency of defective vpu compared with tat and rev genes in brain from patients with HIV type 1-associated dementia. AIDS Res Hum Retroviruses 2007; 23:575-80; PMID:17451348; http://dx.doi.org/10.1089/aid.2006.0246
- Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molcular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses 1996; 12:191-4; PMID:8835195; http://dx.doi.org/10.1089/aid.1996.12.191
- Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol 1991; 65:3973-85; PMID:1830110
- Bannert N, Schenten D, Craig S, Sodroski J. The Level of CD4 Expression Limits Infection of Primary Rhesus Monkey Macrophages by a T-Tropic Simian Immunodeficiency Virus and Macrophagetropic Human Immunodeficiency Viruses. J Virol 2000; 74:10984-93; PMID:11069993; http://dx.doi.org/10.1128/JVI.74.23.10984-10993.2000
- Jolly C, Booth NJ, Neil SJD. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J Virol 2010; 84:12185-99; PMID:20861257; http://dx.doi.org/10.1128/JVI.01447-10
- Chan JK, Greene WC. Dynamic roles for NF-κB in HTLV-I and HIV-1 retroviral pathogenesis. Immunol Rev 2012; 246:286-310; PMID:22435562; http://dx.doi.org/10.1111/j.1600-065X.2012.01094.x
- Krummheuer J, Johnson AT, Hauber I, Kammler S, Anderson JL, Hauber J, Purcell DFJ, Schaal H. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology 2007; 363:261-71; PMID:17331561; http://dx.doi.org/10.1016/j.virol.2007.01.022
- Langer SM, Hopfensperger K, Iyer SS, Kreider EF, Learn GH, Lee L-H, Hahn BH, Sauter D. A Naturally Occurring rev1-vpu Fusion Gene Does Not Confer a Fitness Advantage to HIV-1. PLOS One 2015; 10:e0142118; PMID:26554585; http://dx.doi.org/10.1371/journal.pone.0142118
- Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci USA 2013; 110:6626-33; PMID:23542380; http://dx.doi.org/10.1073/pnas.1304288110
- Asmal M, Hellmann I, Liu W, Keele BF, Perelson AS, Bhattacharya T, Gnanakaran S, Daniels M, Haynes BF, Korber BT, et al. A signature in HIV-1 envelope leader peptide associated with transition from acute to chronic infection impacts envelope processing and infectivity. PLoS ONE 2011; 6:e23673; PMID:21876761; http://dx.doi.org/10.1371/journal.pone.0023673
- Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, Shen T, Gaschen B, Krishnamoorthy M, Li H, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog 2011; 7:e1002209; PMID:21980282; http://dx.doi.org/10.1371/journal.ppat.1002209
- Malim MH, Emerman M. HIV-1 Accessory Proteins—Ensuring Viral Survival in a Hostile Environment. Cell Host & Microbe 2008; 3:388-98; PMID:18541215; http://dx.doi.org/10.1016/j.chom.2008.04.008
- Mashiba M, Collins DR, Terry VH, Collins KL. Vpr overcomes macrophage-specific restriction of HIV-1 Env expression and virion production. Cell Host Microbe 2014; 16:722-35; PMID:25464830; http://dx.doi.org/10.1016/j.chom.2014.10.014
- Laguette N, Brégnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 2014; 156:134-45; PMID:24412650; http://dx.doi.org/10.1016/j.cell.2013.12.011
