ABSTRACT
Members of the ADP-ribosylation factor (Arf) family of small GTP-binding (G) proteins regulate several aspects of membrane trafficking, such as vesicle budding, tethering and cytoskeleton organization. Arf family members, including Arf-like (Arl) proteins have been implicated in several essential cellular functions, like cell spreading and migration. These functions are used by cancer cells to disseminate and invade the tissues surrounding the primary tumor, leading to the formation of metastases. Indeed, Arf and Arl proteins, as well as their guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) have been found to be abnormally expressed in different cancer cell types and human cancers. Here, we review the current evidence supporting the involvement of Arf family proteins and their GEFs and GAPs in cancer progression, focusing on 3 different mechanisms: cell-cell adhesion, integrin internalization and recycling, and actin cytoskeleton remodeling.
Introduction
The ADP-ribosylation factor (Arf) family of small GTP-binding (G) proteins belongs to the Ras superfamily, whose members regulate all aspects of membrane traffic. In particular, Arf family members regulate vesicle budding, tethering and cytoskeleton organization.Citation1 Mammals express 6 Arf isoforms, Arf1-6, which are classified according to their sequence homology in class I (Arf1, 2 and 3), class II (Arf4 and 5) and class III (Arf6).Citation1 The Arf-like (Arl) proteins also belong to the Arf family and show a greater diversity, with 22 members in humans.Citation2 Similar to other members of the Ras superfamily, Arfs and Arls alternate between a GTP-bound active and a GDP-bound inactive state. The exchange of GDP for GTP is catalyzed by guanine nucleotide exchange factors (GEFs) and the inactivation of these proteins requires GTPase-activating proteins (GAPs). Arf proteins associate with membranes generally via myristoylation, only when they are active.Citation3
Arf family proteins are involved in several essential functions, such as cell adhesion and migration. Interestingly, cancer cells subvert these functions in order to disseminate. Indeed, tumor cells can colonize distant organs and form metastases, a process that requires cell migration from the primary tumor and invasion of the surrounding tissues. The acquisition by tumor cells of migratory and invasive capacities results primarily from the loss of apical-basal polarity and cell-cell contacts. Additionally, to migrate through the surrounding extracellular matrix (ECM), tumor cells need to secrete hydrolytic enzymes to degrade it. Furthermore, cell migration and invasion require the internalization and recycling of integrins and the remodeling of the actin cytoskeleton.Citation4 The actin cytoskeleton and actin-binding proteins control the dynamics of cell-cell and cell-ECM contacts and the formation of polarized actin-based migratory protrusions, such as lamellipodia, filopodia, apically-restricted circular dorsal ruffles (CDRs) and invadopodia.Citation5
Roles for Arf/Arl proteins in cancer progression have recently emerged. Indeed, aberrant activity or expression of members of this family has been described as playing a role in cancer cell migration, invasion and proliferation. In particular, Arf1, Arf4 and Arf6 are abnormally expressed in different cancer cell types and human cancers, such as breast, gastric, prostate and lung (). Moreover, the expression of the Arl protein Arl4 was found to be upregulated in leiomyosarcoma subtype II, colon and lung cancers and downregulated in ovarian cancer (). Not surprisingly, Arf GEFs and GAPs have also emerged as candidate regulators of cancer progression. Indeed, their expression was found altered in distinct types of cancers like breast, colon, lung, ovarian and hepatocellular carcinomas (). Furthermore, their expression has been correlated with higher grade tumors, suggesting a crucial role for these regulators of Arf activity in cancer.
Table 1. Expression of Arf family proteins in neoplastic tissues and cancer cells. (+) upregulated; (−) downregulated.
Table 2. Expression of Arf guanine nucleotide exchange factors in neoplastic tissues and cancer cells. (+) upregulated; (−) downregulated.
Table 3. Expression of Arf GTPase-activating proteins in neoplastic tissues and cancer cells. (+) upregulated; (−) downregulated.
In this mini-review, we summarize the current knowledge of the roles that Arf/Arl proteins and their GEFs and GAPs play in cancer cell migration and invasiveness. For this, we chose to focus on 3 processes that are involved in these functions, namely cell-cell adhesion, integrin internalization and recycling, and actin cytoskeleton remodeling.
Regulation of cell adhesion in cancer cells by Arf family proteins
In order to initiate migration, cancer cells must detach from the primary tumor site by disassembling their cell-cell contacts and establishing new ones with the ECM. The adhesion of epithelial cells is mediated by distinct multiprotein complexes, namely adherens junctions (AJs), tight junctions (TJs) and focal adhesions (FAs). Arfs and their GEFs/GAPs have emerged as modulators of these complexes. While Arf6 and one of its GEFs and GAPs are prominently involved in the regulation of cell-cell adhesion through AJ formation and turnover, the interaction of Arf1 and several Arf GAPs with FA components regulates cell-ECM adhesion.
E-cadherin is the best studied component of AJs and the major factor required for cell-cell adhesion. Arf1 regulates cell-cell adhesion by repressing the formation of AJs in an E-cadherin-dependent manner ().Citation6 Arf6 has been implicated in both the assembly and the disassembly of AJs ().Citation1 Moreover, Arf6 regulates E-cadherin internalization into early endosomes and trafficking between the basolateral plasma membrane and early endosomes, facilitating the disassembly of AJs.Citation7 The cycling of Arf6 between active and inactive states, through the action of its GEFs and GAPs seems to be critical for AJ turnover. Indeed, E-cadherin-mediated cell-cell adhesion is impaired in poorly invasive breast cancer MCF7 cells overexpressing the Arf6 GEF BRAG2, when stimulated with epidermal growth factor (EGF).Citation8 This leads to the acquisition of a more invasive phenotype by these cells. On the other hand, BRAG2 silencing leads to increased levels of E-cadherin at the cell surface in pancreatic cancer cells and defects in E-cadherin internalization induced by hepatocyte growth factor (HGF), in cervical carcinoma cells.Citation9,10 Additionally, upon establishment of E-cadherin-mediated cell-cell contacts, the Arf6 GEF EFA6 regulates the de novo formation of TJs in Madin-Darby canine kidney (MDCK) cells, requiring its GEF and actin remodeling activities ().Citation11 Concerning the role of Arf GAPs in E-cadherin trafficking, the overexpression of the Arf6 GAP SMAP1 strongly inhibits clathrin-dependent endocytosis of E-cadherin, leading to a decrease in cell migration and spreading.Citation12 A similar effect on E-cadherin endocytosis was observed with a GAP-negative mutant of SMAP1.Citation12
Figure 1. Schematic diagram illustrating the role of Arf family proteins and their regulators in cancer cell migration. (A and B) Regulation of cell-cell and cell-ECM adhesion by Arf family proteins and their GEFs/GAPs. The initial steps of cancer cell migration involve the disassembly of cell-cell contacts and the establishment of new focal contacts. These are mediated by distinct multiprotein complexes that form adherens and tight junctions, as well as focal adhesions. (A) Members of the Arf family of proteins and their regulators involved in the internalization and recycling of E-cadherin, the best studied component of adherens junctions, and tight junction formation. (B) Arf1 and several Arf GAPs have been shown to regulate focal adhesion formation and turnover. Inset shows the association of Arf1 and Arf GAPs with focal adhesion components. (C) Arfs/Arls and their respective GEFs/GAPs involved in integrin endocytosis and recycling, and actin cytoskeleton remodeling, through the formation of lamellipodia and circular dorsal ruffles. Inset shows Arfs and Arf GEFs and GAPs that regulate the formation of lamellipodia and actin stress fibers. Arfs/Arls are represented in orange ovals. GEFs are represented in green rectangles and GAPs in pink rectangles. CDR, circular dorsal ruffle; EE, early endosome; LE, late endosome; MP, macropinosome; RE, recycling endosome.
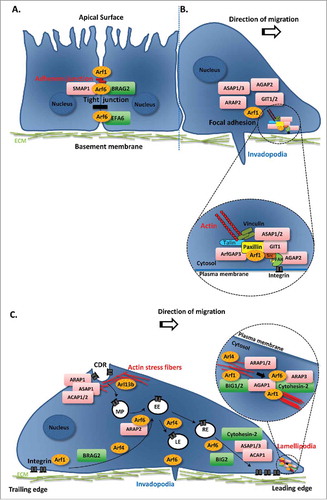
FAs are composed of protein complexes that include among others, kinases, adaptors and actin-binding proteins, which together couple the cell membrane to the actin cytoskeleton and the ECM.Citation13 Arf1 was shown to regulate the formation of FAs in invasive breast cancer cells through the phosphorylation of FA kinase (FAK). Indeed, Arf1 interacts with several proteins within the FA complex, like paxillin, talin and FAK, regulating their interactions (, inset).Citation14 Furthermore, several Arf GAPs have been found to associate with FAs, including ASAP1, ASAP3, GIT1, GIT2, ARAP2 and AGAP2 ().Citation15-Citation20 Overexpression of ASAP1, ASAP2 or GIT1 is associated with a reduction of paxillin and/or FAK at FAs, leading to the inhibition of cell spreading,Citation15 mediated by ASAP1 GAP activity and altered cell motility.Citation17,21 In contrast, overexpression of ArfGAP3 leads to an increase in the levels of paxillin in FAs, which is associated with an increase in cell migration of prostate cancer cells.Citation22 Regulation of FAs through the binding to FAK was also observed for AGAP2, since its silencing leads to a decrease in FAK activity upon stimulation with EGF or platelet-derived growth factor (PDGF). This results in FA stabilization and changes in cell morphology, as well as a decrease in cell migration.Citation20 In agreement, AGAP2 overexpression results in FA dissolution, which is independent of its GAP activity.Citation20 Moreover, GIT1 overexpression results in the disassembly of focal complexes,Citation17 while its silencing inhibits cell migration and leads to a decrease in the levels of FA components like paxillin, phospho-paxillin and FAK.Citation23,24
Arf family proteins in integrin internalization and recycling
Actively migrating and invading cells display an increased requirement for internalization and recycling of integrins, to promote the disassembly of FAs and the detachment of the cell from the ECM at the trailing edge and the establishment of new cell-ECM contacts at the leading edge. Importantly, integrin-mediated cell adhesion and migration have been associated with tumor development.Citation25 Since several Arfs like Arf1, Arf4 and Arf6 regulate integrin trafficking, it is not surprising that they have an impact on the migratory and invasive capacities of cancer cells. Indeed, it has been demonstrated that Arf1 controls cell migration and proliferation of breast cancer cells.Citation26 Arf1 interacts with β1-integrin and regulates the interaction between this integrin and key proteins of FAs, such as paxillin, talin and FAK ().Citation14 Moreover, Arf4 has been shown to control the internalization of α5β1-integrins, regulating their trafficking to late endosomes/lysosomes and consequently their degradation.Citation27 Furthermore, Arf6 has been shown to be required for cell spreading through β1-integrin recycling, since the expression of a dominant-negative form of Arf6 inhibits the recycling of this integrin to the plasma membrane ().Citation28
The role of these Arf proteins in integrin trafficking and consequently on cell migration may be explained by the action of distinct Arf GEFs and GAPs. In fact, BRAG2 regulates cell adhesion and spreading by controlling β1-integrin levels at the cell surface ().Citation29 Moreover, the depletion of BIG1 or BIG2, which belong to a subfamily of Arf1 and Arf3 GEFs leads to a decrease in cancer cell migration. This effect could be explained by impaired β1-integrin N-linked glycosylation, in the case of BIG1,Citation30 and β1-integrin mislocalization to the perinuclear area, in the case of BIG2.Citation31 Additionally, the Arf GEF cytohesin family has been implicated in the regulation of cell migration through the control of integrin internalization and recycling. Overexpression of cytohesin-1 induces αLβ2 integrin-mediated spreading and adhesion, through the interaction with the cytoplasmic domain of the β2-integrin chain.Citation32,33 Cytohesin-2 (a.k.a. Arf nucleotide-binding site opener or ARNO) positively regulates cell migration since its overexpression leads to an increase in the cell migratory ability,Citation34 whereas its downregulation leads to a decrease in cell migration.Citation35,36 In contrast, cytohesin-3 is a negative regulator of cell migration, since its silencing enhances cell adhesion, spreading, and migration.Citation36 These opposite roles in the regulation of cell migration may in part be explained by the differential effect of these 2 members of the cytohesin family on β1-integrin recycling, their distinct cellular localization in spreading cells and/or the interaction with different partners through their coiled-coil domains, which are only 50% identical. Indeed, cytohesin-2 is required for β1-integrin recycling, whereas cytohesin-3 is not.Citation36 Cytohesin-2 was also shown to be required for the serum-induced recycling of α5-integrin, a subunit of the α5β1-integrin heterodimer ().Citation37
Finally, the Arf GAPs ARAP2, ACAP1 and ASAP1/3 have been reported to play a role in integrin trafficking in cancer cells. ARAP2 colocalizes with Arf6 in early endosomes that are positive for APPL1 (adaptor protein phosphotyrosine interacting with PH and leucine zipper 1) and its silencing decreases β1-integrin internalization ().Citation38 In contrast, ACAP1 localizes to tubular recycling endosomes and its silencing enhances integrin internalization and reduces integrin recycling through the slow recycling pathway ().Citation38 Moreover, phosphorylation of ACAP1 by protein kinase B (PKB, a.k.a. Akt) is essential for its association with β1-integrin and β1-integrin recycling, during cell migration.Citation39 Furthermore, ASAP1/3 silencing has been shown to decrease β1-integrin recycling to the plasma membrane ().Citation40,41
Arf-mediated regulation of growth factor-stimulated actin cytoskeleton remodeling
Reorganization of the actin cytoskeleton is crucial for the migratory and invasive behavior of cancer cells. As cancer cells migrate, intracellular signaling cascades are activated to promote remodeling of the actin cytoskeleton and form membrane protrusive structures like lamellipodia and invadopodia.Citation5 Additionally, growth factor stimulation triggers the formation of highly dynamic circular dorsal ruffles (CDRs), which propagate waves along the dorsal plasma membrane.Citation5,42 Although the function of CDRs remains poorly defined, especially in cancer, these structures are thought to have a role in the fast disassembly and remodeling of filamentous actin prior to lamellipodia formation. Moreover, they are involved in directed cell motility and internalization of receptor tyrosine kinases (RTKs) and other membrane receptors.Citation42 Members of the Arf family have been characterized as key players in regulating actin remodeling. Indeed, both Arf1 and Arf6 induce alterations in the actin cytoskeleton through the regulation of the trafficking of Rac1, a small GTPase known to regulate cancer cell migration. For instance, Arf1 silencing impairs the ability of Rac1 to interact with the insulin receptor substrate (IRSp53), an essential step to control actin nucleation and lamellipodia formation ().Citation43 Additionally, Arf1 has been shown to regulate migration and invasion of breast cancer cells through the Rho GTPases RhoA and RhoC, which are required for the phosphorylation of myosin regulatory light-chain (RLC).Citation44 On the other hand, Arf6 can induce Rac1 trafficking to the plasma membrane and regulate Rac1 activation.Citation45 Moreover, in MDA-MB-231 breast cancer cells Arf6 has been implicated in the formation of invadopodia, which are specialized structures formed at the ventral surface of cells that constitute sites for degradation of the ECM ().Citation46
Arl proteins have been also shown to modulate actin cytoskeleton remodeling. The three Arl4 isoforms, Arl4a, c and d can recruit cytohesin-2 to the plasma membrane.Citation47 Arl4a associates with the engulfment and cell motility (ELMO) proteins to promote actin cytoskeleton remodeling.Citation48 Silencing of Arl4c inhibits Rac1 activity, leading to a decrease in cell migration, invasion and proliferation of colorectal and lung cancer cells.Citation49 Arl4d is an upstream regulator of cytohesin-2 that promotes Arf6 activation and also modulates actin remodeling. Indeed, an active form of Arl4d increases Arf6-GTP and induces the disassembly of actin stress fibers.Citation50 Another Arl protein, Arl13b was recently associated by us with actin remodeling and cell migration. Arl13b has been prominently associated with ciliogenesis and sonic hedgehog signaling,Citation51,52 which plays a critical role in aggressive cancers.Citation53 We have found that Arl13b interacts with the non-muscle myosin IIA (NMIIA) in a GTP-dependent manner and regulates CDR formation and fibroblast migration ().Citation54 Additionally, our unpublished results indicate that Arl13b regulates cancer cell progression since its silencing leads to the impairment of breast and colon cancer cell migration and invasion. In contrast, Arl13b overexpression enhances cancer cell migration and invasion and its expression is found upregulated in highly invasive breast cancer cell lines (Casalou C., Faustino A. and Barral D.C., unpublished results).
Arf GEFs and GAPs have also emerged as regulators of actin cytoskeleton remodeling, a process subverted by cancer cells to spread and invade. Cytohesin-2 has been described to play a role in plasma membrane ruffling induced by the calcium-sensing receptor (CaSR) ().Citation55 Cytohesin-2-induced lamellipodia formation and cell motility requires the activation of Rac1, which is mediated by the dedicator of cytokinesis 180/ELMO complex (Dock180).Citation56-Citation58 BIG1 silencing leads to a decrease in cell motility and is associated with defects in orientation of Golgi/microtubule-organizing center (MTOC).Citation59 Moreover, depletion of BIG2 leads to a reduction in actin-dependent membrane protrusions and actin-binding proteins (Arp2, Arp3, cofilin and phosphorylated cofilin) at the leading edge of migrating cells.Citation31 Furthermore, silencing of BIG1 or BIG2 results in an increase in the phosphorylation of the RLC of NMIIA, leading to an enhanced content in actin stress fibers and impaired cell migration, independently of the Arf GEF activity ().Citation60 Finally, the requirement for BRAG2 in EGF receptor (EGFR)-induced cell migration and activation of extracellular signal-regulated kinase (ERK)/Rac1 signaling has been shown in hepatoma and breast cancer cells.Citation61,62
The Arf GAPs ARAP1/2/3, AGAP1, ASAP1 and ACAP1/2 have also been found to regulate actin-rich membrane ruffles. Indeed, ARAP1 overexpression, which preferentially inactivates Arf1 and Arf5, leads to cell rounding and loss of actin stress fibers, mediated by its Rho GAP domain and resulting in inhibition of cell spreading.Citation63 Furthermore, it has been shown that ARAP1 expression leads to an increase in the size of the ring structure of CDRs in an Arf GAP activity-dependent manner ().Citation64 In the case of ARAP2, which selectively uses Arf6 as a substrate, its silencing leads to a decrease in the formation of FAs and actin stress fibers, in an Arf GAP activity- and RhoA-GTP binding-dependent manner ().Citation65 ARAP2 also regulates Rac1 activity, since its silencing increases the levels of the activated form of Rac1.Citation19 ARAP3 overexpression leads to the formation of membrane projections and to a decrease in cell spreading and cell migration.Citation66 These phenotypes are associated with reduced levels of RhoA and Rac1 and independent of the Arf GAP domain.Citation66 The overexpression of AGAP1, which inactivates preferentially Arf1 and Arf5, induces the formation of lamellipodia ()Citation67 and the loss of stress fibers and PDGF-induced ruffles.Citation68 ASAP1 was reported to act as a positive regulator of cell motility/migration in some studies,Citation69-Citation72 while another study reported that it acts as a negative regulator of these processes.Citation73 ASAP1 localizes to CDRs and negatively regulates their number upon PDGF stimulation. A similar effect on CDRs was observed by overexpressing the Arf GAPs ACAP1 or ACAP2 ().Citation74
Conclusions and perspectives
In this mini-review we described the evidence supporting the involvement of Arf/Arl proteins and their GEFs and GAPs in cancer cell migration and invasion through several mechanisms, including the regulated trafficking of adhesion molecules and integrins and rearrangement of the actin cytoskeleton (). In light of this, Arf family proteins and their regulators constitute putative targets for novel anti-cancer therapies. Arf proteins fulfill many fundamental roles besides the ones reviewed here. Therefore, Arf GEFs and GAPs are more attractive to serve as therapeutic targets. Indeed, several studies in mouse models show that this is a promising strategy. AMF-26, an inhibitor of Arf1 activation, was shown to induce the regression of human breast cancer xenografts.Citation75 Moreover, the cytohesin family inhibitor SecinH3 suppresses melanoma growth and angiogenesis.Citation76 Furthermore, an analog of the phosphatidylinositol-3,4,5-trisphosphate antagonist PIT-1, which inhibits Arf6 activation was shown to impair the formation of pulmonary metastases upon melanoma cell injection.Citation77 Nevertheless, a better understanding of the molecular mechanisms of membrane trafficking regulated by Arf small G proteins is needed to understand the critical roles that these proteins have in several biological processes, including cancer progression, as well as to discover novel anti-cancer strategies.
Abbreviations
| AJ | = | adherens junction |
| Akt | = | protein kinase B |
| APPL1 | = | adaptor protein phosphotyrosine interacting with PH and leucine zipper 1 |
| Arf | = | ADP-ribosylation factor |
| Arl | = | Arf-like |
| ARNO | = | Arf nucleotide-binding site opener |
| CaSR | = | calcium-sensing receptor |
| CDR | = | circular dorsal ruffle |
| Dock180 | = | dedicator of cytokinesis of 180 kDa |
| ECM | = | extracellular matrix |
| EGF | = | epidermal growth factor |
| EGFR | = | epidermal growth factor receptor |
| ELMO | = | engulfment and cell motility |
| ERK | = | extracellular signal-regulated kinase |
| FA | = | focal adhesion |
| FAK | = | focal adhesion kinase |
| GAP | = | GTPase-activating protein |
| GEF | = | guanine nucleotide exchange factor |
| HGF | = | hepatocyte growth factor |
| IRSp53 | = | insulin receptor substrate of 53 kDa |
| MDCK | = | Madin-Darby canine kidney |
| MTOC | = | microtubule-organizing center |
| NMIIA | = | non-muscle myosin IIA |
| PDGF | = | platelet-derived growth factor |
| PDGFR | = | platelet-derived growth factor receptor |
| RLC | = | regulatory light chain |
| RTK | = | receptor tyrosine kinase |
| TJ | = | tight junction |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank members of the Membrane Traffic in Infection and Disease lab for helpful discussions. We apologize to those whose work could not be included because of space limitations.
Funding
C.C. is funded by a postdoctoral fellowship from Fundação para a Ciência e Tecnologia (FCT; SFRH/BPD/78561/2011), A.F. by a PhD fellowship from FCT (PD/BD/105898/2014) and D.C.B. by the Investigator FCT program (IF/00501/2014/CP1252/CT0001). Our work is supported by FCT (PTDC/BIM-MEC/4905/2014 and iNOVA4Health - UID/Multi/04462/2013).
References
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 2011; 12:362-75; PMID:21587297; http://dx.doi.org/10.1038/nrm3117
- Gillingham AK, Munro S. The Small G Proteins of the Arf Family and Their Regulators. Annu Rev Cell Dev Biol 2007; 23:579-611; PMID:17506703; http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123209
- Jackson CL, Bouvet S. Arfs at a glance. J Cell Sci 2014; 127:4103-9; PMID:25146395; http://dx.doi.org/10.1242/jcs.144899
- Paul NR, Jacquemet G, Caswell PT. Endocytic Trafficking of Integrins in Cell Migration. Curr Biol 2015; 25:R1092-105; PMID:26583903; http://dx.doi.org/10.1016/j.cub.2015.09.049
- Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol 2004; 5:647-57; PMID:15366708; http://dx.doi.org/10.1038/nrm1436
- Schlienger S, Campbell S, Pasquin S, Gaboury L, Claing A, Schlienger S, Campbell S, Pasquin S, Gaboury L, Claing A. ADP-ribosylation factor 1 expression regulates epithelial-mesenchymal transition and predicts poor clinical outcome in triple-negative breast cancer. Oncotarget 2016; 7:15811-27; PMID:26908458
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 2006; 7:347-58; PMID:16633337; http://dx.doi.org/10.1038/nrm1910
- Morishige M, Hashimoto S, Ogawa E, Toda Y, Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat Cell Biol 2008; 10:85-92; PMID:18084281; http://dx.doi.org/10.1038/ncb1672
- Hiroi T, Someya A, Thompson W, Moss J, Vaughan M. GEP100/BRAG2: activator of ADP-ribosylation factor 6 for regulation of cell adhesion and actin cytoskeleton via E-cadherin and alpha-catenin. Proc Natl Acad Sci U S A 2006; 103:10672-7; PMID:16807291; http://dx.doi.org/10.1073/pnas.0604091103
- Xie C, Wei S, Chen J, Xu X, Cai J, Chen Q, Jia L, Li J, Wientjes M, Au J, et al. Down-regulation of GEP100 causes increase in E-cadherin levels and inhibits pancreatic cancer cell invasion. PLoS One 2012; 7:e37854; PMID:22662237; http://dx.doi.org/10.1371/journal.pone.0037854
- Luton F, Klein S, Chauvin J-P, Bivic A Le, Bourgoin S, Franco M, Chardin P. EFA6, exchange factor for ARF6, regulates the actin cytoskeleton and associated tight junction in response to E-cadherin engagement. Mol Biol Cell 2004; 15:1134-45; PMID:14668475; http://dx.doi.org/10.1091/mbc.E03-10-0751
- Kon S, Tanabe K, Watanabe T, Sabe H, Satake M. Clathrin dependent endocytosis of E-cadherin is regulated by the Arf6GAP isoform SMAP1. Exp Cell Res 2008; 314:1415-28; PMID:18331728; http://dx.doi.org/10.1016/j.yexcr.2007.11.006
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta 2004; 1692:103-19; PMID:15246682; http://dx.doi.org/10.1016/j.bbamcr.2004.04.007
- Schlienger S, Ramirez RAM, Claing A. ARF1 regulates adhesion of MDA-MB-231 invasive breast cancer cells through formation of focal adhesions. Cell Signal 2015; 27:403-15; PMID:25530216; http://dx.doi.org/10.1016/j.cellsig.2014.11.032
- Liu Y, Loijens JC, Martin KH, Karginov AV, Parsons JT. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol Biol Cell 2002; 13:2147-56; PMID:12058076; http://dx.doi.org/10.1091/mbc.E02-01-0018
- Ha VL, Bharti S, Inoue H, Vass WC, Campa F, Nie Z, de Grammont A, Ward Y, Randazzo PA. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol Chem 2008; 283(22): 14914-26; PMID:18400762; http://dx.doi.org/10.1074/jbc.M709717200
- Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol 2000; 20(17):6354-63; PMID:10938112; http://dx.doi.org/10.1128/MCB.20.17.6354-6363.2000
- Turner CE, Brown MC, Perrotta JA, Riedy MC, Nikolopoulos SN, McDonald AR, Bagrodia S, Thomas S, Leventhal PS. Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: A role in cytoskeletal remodeling. J Cell Biol 1999; 145(4): 851-63; PMID:10330411; http://dx.doi.org/10.1083/jcb.145.4.851
- Chen PW, Jian X, Yoon HY, Randazzo PA. ARAP2 signals through Arf6 and Rac1 to control focal adhesion morphology. J Biol Chem 2013; 288(8): 5849-60; PMID:23295182; http://dx.doi.org/10.1074/jbc.M112.415778
- Zhu Y, Wu Y, Kim JI, Wang Z, Daaka Y, Nie Z. Arf GTPase-activating protein AGAP2 regulates focal adhesion kinase activity and focal adhesion remodeling. J Biol Chem 2009; 284:13489-96; PMID:19318351; http://dx.doi.org/10.1074/jbc.M900469200
- Kondo A, Hashimoto S, Yano H, Nagayama K, Mazaki Y, Sabe H. A new paxillin-binding protein, PAG3/Papalpha/KIAA0400, bearing an ADP-ribosylation factor GTPase-activating protein activity, is involved in paxillin recruitment to focal adhesions and cell migration. Mol Biol Cell 2000; 11(4):1315-27; PMID:10749932; http://dx.doi.org/10.1091/mbc.11.4.1315
- Obinata D, Takayama K, Urano T, Murata T, Ikeda K, Horie-Inoue K, Ouchi Y, Takahashi S, Inoue S. ARFGAP3, an androgen target gene, promotes prostate cancer cell proliferation and migration. Int J Cancer 2012; 130:2240-8; PMID:21647875; http://dx.doi.org/10.1002/ijc.26224
- Huang W-C, Chan S-H, Jang T-H, Chang J-W, Ko Y-C, Yen T-C, Chiang S-L, Chiang W-F, Shieh T-Y, Liao C-T, et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res 2014; 74:751-64; PMID:24335959; http://dx.doi.org/10.1158/0008-5472.CAN-13-1297
- Chan S-H, Huang W-C, Chang J-W, Chang K-J, Kuo W-H, Wang M-Y, Lin K-Y, Uen Y-H, Hou M-F, Lin C-M, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene 2014; 33:4496-507; PMID:24608434; http://dx.doi.org/10.1038/onc.2014.10
- De Franceschi N, Hamidi H, Alanko J, Sahgal P, Ivaska J. Integrin traffic - the update. J Cell Sci 2015; 128:839-52; PMID:25663697; http://dx.doi.org/10.1242/jcs.161653
- Boulay P-L, Schlienger S, Lewis-Saravalli S, Vitale N, Ferbeyre G, Claing A. ARF1 controls proliferation of breast cancer cells by regulating the retinoblastoma protein. Oncogene 2011; 30:3846-61; PMID:21478909; http://dx.doi.org/10.1038/onc.2011.100
- Rainero E, Howe JD, Caswell PT, Jamieson NB, Anderson K, Critchley DR, Machesky L, Norman JC. Ligand-Occupied Integrin Internalization Links Nutrient Signaling to Invasive Migration. Cell Rep 2015; 10:398-413; PMID:25600874; http://dx.doi.org/10.1016/j.celrep.2014.12.037
- Powelka AM, Sun J, Li J, Gao M, Shaw LM, Sonnenberg A, Hsu VW. Stimulation-dependent recycling of integrin beta1 regulated by ARF6 and Rab11. Traffic 2004; 5:20-36; PMID:14675422; http://dx.doi.org/10.1111/j.1600-0854.2004.00150.x
- Dunphy JL, Moravec R, Ly K, Lasell TK, Melancon P, Casanova JE. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr Biol 2006; 16:315-20; PMID:16461286; http://dx.doi.org/10.1016/j.cub.2005.12.032
- Shen X, Hong M-S, Moss J, Vaughan M. BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein, is required for correct glycosylation and function of integrin beta1. Proc Natl Acad Sci U S A 2007; 104:1230-5; PMID:17227842; http://dx.doi.org/10.1073/pnas.0610535104
- Shen X, Li CC, Aponte AM, Shen R-F, Billings E, Mossa J, Vaughana M. Brefeldin A-inhibited ADP-ribosylation factor activator BIG2 regulates cell migration via integrin beta1 cycling and actin remodeling. Proc Natl Acad Sci U S A 2012; 109:14464-9; PMID:22908276; http://dx.doi.org/10.1073/pnas.1211877109
- Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. Alpha L beta 2 integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell 1996; 86:233-42; PMID:8706128; http://dx.doi.org/10.1016/S0092-8674(00)80095-1
- Geiger C, Nagel W, Boehm T, Kooyk Y van, Figdor CG, Kremmer E, Hogg N, Zeitlmann L, Dierks H, Weber KSC, et al. Cytohesin-1 regulates beta-2 integrin-mediated adhesion through both ARF-GEF function and interaction with LFA-1. EMBO J 2000; 19:2525-36; PMID:10835351; http://dx.doi.org/10.1093/emboj/19.11.2525
- Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol 2001; 154:599-610; PMID:11481345; http://dx.doi.org/10.1083/jcb.200104019
- Pan T, Sun J, Hu J, Hu Y, Zhou J, Chen Z, Xu D, Xu W, Zheng S, Zhang S, et al. Cytohesins/ARNO: the function in colorectal cancer cells. PLoS One 2014; 9:e90997; PMID:24618737; http://dx.doi.org/10.1371/journal.pone.0090997
- Oh SJ, Santy LC. Differential effects of cytohesins 2 and 3 on beta1 integrin recycling. J Biol Chem 2010; 285:14610-6; PMID:20223830; http://dx.doi.org/10.1074/jbc.M109.043935
- Salem JC, Reviriego-Mendoza MM, Santy LC. ARF-GEF cytohesin-2/ARNO regulates R-Ras and alpha5-integrin recycling through an EHD1-positive compartment. Mol Biol Cell 2015; 26:4265-79; PMID:26378252; http://dx.doi.org/10.1091/mbc.E15-05-0278
- Chen PW, Luo R, Jian X, Randazzo PA. The Arf6 GTPase-activating proteins ARAP2 and ACAP1 define distinct endosomal compartments that regulate integrin alpha5beta1 traffic. J Biol Chem 2014; 289:30237-48; PMID:25225293; http://dx.doi.org/10.1074/jbc.M114.596155
- Li J, Ballif BA, Powelka AM, Dai J, Gygi SP, Hsu VW. Phosphorylation of ACAP1 by Akt regulates the stimulation-dependent recycling of integrin beta1 to control cell migration. Dev Cell 2005; 9:663-73; PMID:16256741; http://dx.doi.org/10.1016/j.devcel.2005.09.012
- Yu X, Wang F, Liu H, Adams G, Aikhionbare F, Liu D, Cao X, Fan L, Hu G, Chen Y, et al. ACAP4 protein cooperates with Grb2 protein to orchestrate epidermal growth factor-stimulated integrin beta1 recycling in cell migration. J Biol Chem 2011; 286:43735-47; PMID:22027826; http://dx.doi.org/10.1074/jbc.M111.278770
- Onodera Y, Nam J-M, Hashimoto A, Norman JC, Shirato H, Hashimoto S, Sabe H. Rab5c promotes AMAP1-PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J Cell Biol 2012; 197:983-96; PMID:22734003; http://dx.doi.org/10.1083/jcb.201201065
- Hoon J-L, Wong W-K, Koh C-G. Functions and regulation of circular dorsal ruffles. Mol Cell Biol 2012; 32:4246-57; PMID:22927640; http://dx.doi.org/10.1128/MCB.00551-12
- Lewis-Saravalli S, Campbell S, Claig A. Arf1 controls Rac1 signaling to regulate migration of MDA-MB-231 invasive breast cancer cells. Cell Signaling 2013; 25:1813-9; PMID:23707487; http://dx.doi.org/10.1016/j.cellsig.2013.05.011
- Schlienger S, Campbell S, Claing A. ARF1 regulates the Rho/MLC pathway to control EGF-dependent breast cancer cell invasion. Mol Biol Cell 2014; 25:17-29; PMID:24196838; http://dx.doi.org/10.1091/mbc.E13-06-0335
- Marchesin V, Montagnac G, Chavrier P. ARF6 promotes the formation of Rac1 and WAVE-dependent ventral F-actin rosettes in breast cancer cells in response to epidermal growth factor. PLoS One 2015; 10:e0121747; PMID:25799492; http://dx.doi.org/10.1371/journal.pone.0121747
- Hashimoto S, Onodera Y, Hashimoto AJ, Tanaka T, Hamaguchi M, Yamada A, Sabe H. Requirement of Arf6 in breast cancer invasive activities. Proc Natl Acad Sci U S A 2004; 101:6647-52; PMID:15087504; http://dx.doi.org/10.1073/pnas.0401753101
- Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 Family of Small G Proteins Can Recruit the Cytohesin Arf6 Exchange Factors to the Plasma Membrane. Curr Biol 2007; 17: 711-6; PMID:17398095; http://dx.doi.org/10.1016/j.cub.2007.03.007
- Patel M, Chiang T-C, Tran V, Lee F-JS, Côté J-F. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J Biol Chem 2011; 286:38969-79; PMID:21930703; http://dx.doi.org/10.1074/jbc.M111.274191
- Fujii S, Matsumoto S, Nojima S, Morii E, Kikuchi A. Arl4c expression in colorectal and lung cancers promotes tumorigenesis and may represent a novel therapeutic target. Oncogene 2015; 34:4834-44; PMID:25486429; http://dx.doi.org/10.1038/onc.2014.402
- Li C-C, Chiang T-C, Wu T-S, Pacheco-Rodriguez G, Moss J, Lee F-JS. ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol Biol Cell 2007; 18:4420-37; PMID:17804820; http://dx.doi.org/10.1091/mbc.E07-02-0149
- Caspary T, Larkins CE, Anderson K V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 2007; 12:767-78; PMID:17488627; http://dx.doi.org/10.1016/j.devcel.2007.03.004
- Larkins CE, Aviles GDG, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 2011; 22:4694-703; PMID:21976698; http://dx.doi.org/10.1091/mbc.E10-12-0994
- Hui M, Cazet A, Nair R, Watkins DN, O'Toole SA, Swarbrick A. The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy. Breast Cancer Res 2013; 15:203; PMID:23547970; http://dx.doi.org/10.1186/bcr3401
- Casalou C, Seixas C, Portelinha A, Pintado P, Barros M, Ramalho JS, Lopes SS, Barral DC. Arl13b and the non-muscle myosin heavy chain IIA are required for circular dorsal ruffle formation and cell migration. J Cell Sci 2014; 127:2709-22; PMID:24777479; http://dx.doi.org/10.1242/jcs.143446
- Bouschet T, Martin S, Kanamarlapudi V, Mundell S, Henley JM. The calcium-sensing receptor changes cell shape via a beta-arrestin-1 ARNO ARF6 ELMO protein network. J Cell Sci 2007; 120:2489-97; PMID:17623778; http://dx.doi.org/10.1242/jcs.03469
- Frank SR, Hatfield JC, Casanova JE. Remodeling of the actin cytoskeleton is coordinately regulated by protein kinase C and the ADP-ribosylation factor nucleotide exchange factor ARNO. Mol Biol Cell 1998; 9:3133-46; PMID:9802902; http://dx.doi.org/10.1091/mbc.9.11.3133
- Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol 2005; 15:1749-54; PMID:16213822; http://dx.doi.org/10.1016/j.cub.2005.08.052
- White DT, McShea KM, Attar MA, Santy LC. GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180. Mol Biol Cell 2010; 21:562-71; PMID:20016009; http://dx.doi.org/10.1091/mbc.E09-03-0217
- Li CC, Kuo JC, Waterman CM, Kiyama R, Moss J, Vaughan M. Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc Natl Acad Sci U S A 2011; 108:19228-33; PMID:22084092; http://dx.doi.org/10.1073/pnas.1117011108
- Le K, Li CC, Ye G, Moss J, Vaughan M. Arf guanine nucleotide-exchange factors BIG1 and BIG2 regulate nonmuscle myosin IIA activity by anchoring myosin phosphatase complex. Proc Natl Acad Sci U S A 2013; 110:E3162-70; PMID:23918382; http://dx.doi.org/10.1073/pnas.1312531110
- Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam J-M, Miura K, Yano H, Onodera Y. The EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis. Traffic 2009; 10:982-93; PMID:19416474; http://dx.doi.org/10.1111/j.1600-0854.2009.00917.x
- Hu Z, Du J, Yang L, Zhu Y, Yang Y, Zheng D, Someya A, Gu L, Lu X, Gray A, et al. GEP100/Arf6 is required for epidermal growth factor-induced ERK/Rac1 signaling and cell migration in human hepatoma HepG2 cells. PLoS One 2012; 7:e38777; PMID:22701712; http://dx.doi.org/10.1371/journal.pone.0038777
- Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, Resau J, Zheng Y, Randazzo PA. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell 2002; 9:109-19; PMID:11804590; http://dx.doi.org/10.1016/S1097-2765(02)00428-8
- Hasegawa J, Tsujita K, Takenawa T, Itoh T. ARAP1 regulates the ring size of circular dorsal ruffles through Arf1 and Arf5. Mol Biol Cell 2012; 23:2481-9; PMID:22573888; http://dx.doi.org/10.1091/mbc.E12-01-0017
- Yoon HY, Miura K, Cuthbert EJ, Davis KK, Ahvazi B, Casanova JE, Randazzo PA. ARAP2 effects on the actin cytoskeleton are dependent on Arf6-specific GTPase-activating-protein activity and binding to RhoA-GTP. J Cell Sci 2006; 119:4650-66; PMID:17077126; http://dx.doi.org/10.1242/jcs.03237
- I STT, Nie Z, Stewart A, Najdovska M, Hall NE, He H, Randazzo PA, Lock P. ARAP3 is transiently tyrosine phosphorylated in cells attaching to fibronectin and inhibits cell spreading in a RhoGAP-dependent manner. J Cell Sci 2004; 117:6071-84; PMID:15546919; http://dx.doi.org/10.1242/jcs.01526
- Xia C, Ma W, Stafford LJ, Liu C, Gong L, Martin JF, Liu M. GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol Cell Biol 2003; 23:2476-88; PMID:12640130; http://dx.doi.org/10.1128/MCB.23.7.2476-2488.2003
- Nie Z, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Takei J, Randazzo PA. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biol Chem 2002; 277:48965-75; PMID:12388557; http://dx.doi.org/10.1074/jbc.M202969200
- Furman C, Short SM, Subramanian RR, Zetter BR, Roberts TM. DEF-1/ASAP1 is a GTPase-activating protein (GAP) for ARF1 that enhances cell motility through a GAP-dependent mechanism. J Biol Chem 2002; 277:7962-9; PMID:11773070; http://dx.doi.org/10.1074/jbc.M109149200
- Ehlers JP, Worley L, Onken MD, Harbour JW. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin Cancer Res 2005; 11:3609-13; PMID:15897555; http://dx.doi.org/10.1158/1078-0432.CCR-04-1941
- Liu Y, Yerushalmi GM, Grigera PR, Parsons JT. Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J Biol Chem 2005; 280:8884-92; PMID:15632162; http://dx.doi.org/10.1074/jbc.M412200200
- Müller T, Stein U, Poletti A, Garzia L, Rothley M, Plaumann D, Thiele W, Bauer M, Galasso A, Schlag P, et al. ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene 2010; 29:2393-403; PMID:20154719; http://dx.doi.org/10.1038/onc.2010.6
- Chen P-W, Jian X, Heissler SM, Le K, Luo R, Jenkins LM, Nagy A, Moss J, Sellers JR, Randazzo PA. The Arf GTPase-activating Protein, ASAP1, Binds Non muscle Myosin 2A to Control Remodeling of the Actomyosin Network. J Biol Chem 2016; 291:7517-26; PMID:26893376; http://dx.doi.org/10.1074/jbc.M115.701292
- Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, Randazzo PA. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol 2000; 151:627-38; PMID:11062263; http://dx.doi.org/10.1083/jcb.151.3.627
- Ohashi Y, Iijima H, Yamaotsu N, Yamazaki K, Sato S, Okamura M, Sugimoto K, Dan S, Hirono S, Yamori T. AMF-26, a Novel Inhibitor of the Golgi System, Targeting ADP-ribosylation Factor 1 (Arf1) with Potential for Cancer Therapy. J Biol Chem 2012; 287:3885-97; PMID:22158626; http://dx.doi.org/10.1074/jbc.M111.316125
- Hongu T, Funakoshi Y, Fukuhara S, Suzuki T, Sakimoto S, Takakura N, Ema M, Takahashi S, Itoh S, Kato M, et al. Arf6 regulates tumour angiogenesis and growth through HGF-induced endothelial β1 integrin recycling. Nat Commun 2015; 6:7925; PMID:26239146; http://dx.doi.org/10.1038/ncomms8925
- Miao B, Skidan I, Yang J, You Z, Fu X, Famulok M, Schaffhausen B, Torchilin V, Yuan J, Degterev A. Inhibition of cell migration by PITENINs: the role of ARF6. Oncogene 2012; 31:4317-32; PMID:22179837; http://dx.doi.org/10.1038/onc.2011.593
- Jang SY, Jang S-W, Ko J. Regulation of ADP-ribosylation factor 4 expression by small leucine zipper protein and involvement in breast cancer cell migration. Cancer Lett 2012; 314:185-97; PMID:22004728; http://dx.doi.org/10.1016/j.canlet.2011.09.028
- Ma W-M, Jiang B, Lai Z-S, Wang X-Y, Geng Y, Han Y-J. Expression and significance of ADP-ribosylation factor 1 (ARF1) in colorectal laterally spreading tumor. Ai Zheng 2005; 24:690-4; PMID:15946480
- Chen X, Su Z, Wang S, Xu H. Clinical and prognostic significance of Arl4c expression in colorectal cancer. Cancer Biomarkers 2016; 16:253-7; PMID:26756615; http://dx.doi.org/10.3233/CBM-150562
- Tsai M-M, Lin PY, Cheng W-L, Tsai C-Y, Chi H-C, Chen C-Y, Tseng Y-H, Cheng Y-F, Chen C-D, Liang Y, et al. Overexpression of ADP-ribosylation factor 1 in human gastric carcinoma and its clinicopathological significance. Cancer Sci 2012; 103:1136-44; PMID:22348287; http://dx.doi.org/10.1111/j.1349-7006.2012.02243.x
- Zhang Y, Du J, Zheng J, Liu J, Xu R, Shen T, Zhu Y, Chang J, Wang H, Zhang Z, et al. EGF-reduced Wnt5a transcription induces epithelial-mesenchymal transition via Arf6-ERK signaling in gastric cancer cells. Oncotarget 2015; 6:7244-61; PMID:25779663; http://dx.doi.org/10.18632/oncotarget.3133
- Masoudi-Nejad A. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS One 2013; 8:e67552; PMID:23874428; http://dx.doi.org/10.1371/journal.pone.0067552
- Oka S, Uramoto H, Shimokawa H, Yamada S, Tanaka F. Epidermal Growth Factor Receptor-GEP100-Arf6 Axis Affects the Prognosis of Lung Adenocarcinoma. Oncology 2014; 86:263-70; PMID:24902879; http://dx.doi.org/10.1159/000360089
- Bani MR, Nicoletti MI, Alkharouf NW, Ghilardi C, Petersen D, Erba E, Sausville EA, Liu ET, Giavazzi R. Gene expression correlating with response to paclitaxel in ovarian carcinoma xenografts. Mol Cancer Ther 2004; 3:111-21; PMID:14985451
- Su D, Katsaros D, Xu S, Xu H, Gao Y, Biglia N, Feng J, Ying L, Zhang P, Benedetto C, et al. ADP-ribosylation factor-like 4C (ARL4C), a novel ovarian cancer metastasis suppressor, identified by integrated genomics. Am J Transl Res 2015; 7:242-56; PMID:25901194
- Taniuchi K, Furihata M, Saibara T. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia 2014; 16:1082-93; PMID:25499221; http://dx.doi.org/10.1016/j.neo.2014.10.007
- Davis JE, Xie X, Guo J, Huang W, Chu WM, Huang S, Teng Y, Wu G. ARF1 promotes prostate tumorigenesis via targeting oncogenic MAPK signaling. Oncotarget 2016; 7:39834-45; PMID:27213581
- Morgan C, Lewis PD, Hopkins L, Burnell S, Kynaston H, Doak SH, Donaldson J, Hashimoto S, Onodera Y, Hashimoto A, et al. Increased expression of ARF GTPases in prostate cancer tissue. Springerplus 2015; 4:342; PMID:26185744; http://dx.doi.org/10.1186/s40064-015-1136-y
- Chi JH, Panner A, Cachola K, Crane CA, Murray J, Pieper RO, James CD, Parsa AT. Increased expression of the glioma-associated antigen ARF4L after loss of the tumor suppressor PTEN. Laboratory investigation. J Neurosurg 2008; 108:299-303; PMID:18240926; http://dx.doi.org/10.3171/JNS/2008/108/2/0299
- Hu B, Shi B, Jarzynka MJ, Yin J-J, D'Souza-Schorey C, Cheng S-Y. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res 2009; 69:794-801; PMID:19155310; http://dx.doi.org/10.1158/0008-5472.CAN-08-2110
- Webster MR, Weeraratna AT, Dunn KJ, Williams BO, Li Y, Pavan WJ, Takeda K, Yasumoto K, Takada R, Takada S, et al. A Wnt-er migration: the confusing role of β-catenin in melanoma metastasis. Sci Signal 2013; 6:pe11; PMID:23532332; http://dx.doi.org/10.1126/scisignal.2004114
- Muralidharan-Chari V, Hoover H, Clancy J, Schweitzer J, Suckow MA, Schroeder V, Castellino FJ, Schorey JS, D'Souza-Schorey C. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res 2009; 69:2201-9; PMID:19276388; http://dx.doi.org/10.1158/0008-5472.CAN-08-1301
- Hongu T, Yamauchi Y, Funakoshi Y, Katagiri N, Ohbayashi N, Kanaho Y. Pathological functions of the small GTPase Arf6 in cancer progression: Tumor angiogenesis and metastasis. Small GTPases 2016; 7:47-53; PMID:26909552; http://dx.doi.org/10.1080/21541248.2016.1154640
- Olstad OK, Gautvik VT, Reppe S, Rian E, Jemtland R, Ohlsson C, Bruland OS, Gautvik KM. Molecular heterogeneity in human osteosarcoma demonstrated by enriched mRNAs isolated by directional tag PCR subtraction cloning. Anticancer Res 2003; 23:2201-16; PMID:12894494
- Kannangai R, Vivekanandan P, Martinez-Murillo F, Choti M, Torbenson M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum Pathol 2007; 38:639-44; PMID:17367606; http://dx.doi.org/10.1016/j.humpath.2006.07.019
- Guo X, Jo VY, Mills AM, Zhu SX, Lee C-H, Espinosa I, Nucci MR, Varma S, Forgó E, Hastie T, et al. Clinically Relevant Molecular Subtypes in Leiomyosarcoma. Clin Cancer Res 2015; 21:3501-11; PMID:25896974; http://dx.doi.org/10.1158/1078-0432.CCR-14-3141
- Kinoshita R, Nam J-M, Ito YM, Hatanaka KC, Hashimoto A, Handa H, Otsuka Y, Hashimoto S, Onodera Y, Hosoda M, et al. Co-overexpression of GEP100 and AMAP1 proteins correlates with rapid local recurrence after breast conservative therapy. PLoS One 2013; 8:e76791; PMID:24116160; http://dx.doi.org/10.1371/journal.pone.0076791
- Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat 2009; 113:239-49; PMID:18293085; http://dx.doi.org/10.1007/s10549-008-9927-2
- Zangari J, Partisani M, Bertucci F, Milanini J, Bidaut G, Berruyer-Pouyet C, Finetti P, Long E, Brau F, Cabaud O, et al. EFA6B antagonizes breast cancer. Cancer Res 2014; 74:5493-506; PMID:25115298; http://dx.doi.org/10.1158/0008-5472.CAN-14-0298
- Wang Y, Han K-J, Pang X-W, Vaughan HA, Qu W, Dong X-Y, Peng J-R, Zhao H-T, Rui J-A, Leng X-S, et al. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol 2002; 169:1102-9; PMID:12097419; http://dx.doi.org/10.4049/jimmunol.169.2.1102
- Menju T, Hashimoto S, Hashimoto A, Otsuka Y, Handa H, Ogawa E, Toda Y, Wada H, Date H, Sabe H. Engagement of overexpressed Her2 with GEP100 induces autonomous invasive activities and provides a biomarker for metastases of lung adenocarcinoma. PLoS One 2011; 6:e25301; PMID:21966491; http://dx.doi.org/10.1371/journal.pone.0025301
- Pils D, Horak P, Gleiss A, Sax C, Fabjani G, Moebus VJ, Zielinski C, Reinthaller A, Zeillinger R, Krainer M. Five genes from chromosomal band 8p22 are significantly down-regulated in ovarian carcinoma: N33 and EFA6R have a potential impact on overall survival. Cancer 2005; 104:2417-29; PMID:16270321; http://dx.doi.org/10.1002/cncr.21538
- van den Boom J, Wolter M, Blaschke B, Knobbe CB, Reifenberger G. Identification of novel genes associated with astrocytoma progression using suppression subtractive hybridization and real-time reverse transcription-polymerase chain reaction. Int J Cancer 2006; 119:2330-8; PMID:16865689; http://dx.doi.org/10.1002/ijc.22108
- Park JY, Kim S-A, Chung JW, Bang S, Park SW, Paik Y-K, Song SY. Proteomic analysis of pancreatic juice for the identification of biomarkers of pancreatic cancer. J Cancer Res Clin Oncol 2011; 137:1229-38; PMID:21691750; http://dx.doi.org/10.1007/s00432-011-0992-2
- Villalva C, Trempat P, Greenland C, Thomas C, Girard JP, Moebius F, Delsol G, Brousset P. Isolation of differentially expressed genes in NPM-ALK-positive anaplastic large cell lymphoma. Br J Haematol 2002; 118:791-8; PMID:12181047; http://dx.doi.org/10.1046/j.1365-2141.2002.03671.x
- Xu K, Gao J, Yang X, Yao Y, Liu Q. Cytohesin-2 as a novel prognostic marker for hepatocellular carcinoma. Oncol Rep 2013; 29:2211-8; PMID:23545718
- Fu Y, Li J, Feng M-X, Yang X-M, Wang Y-H, Zhang Y-L, Qin W, Xia Q, Zhang Z-G. Cytohesin-3 is upregulated in hepatocellular carcinoma and contributes to tumor growth and vascular invasion. Int J Clin Exp Pathol 2014; 7:2123-32; PMID:24966920
- Liu X, Hu Y, Hao C, Rempel SA, Ye K. PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene 2007; 26:4918-27; PMID:17297440; http://dx.doi.org/10.1038/sj.onc.1210290
- Peng H, Dara L, Li TWH, Zheng Y, Yang H, Tomasi ML, Tomasi I, Giordano P, Mato JM, Lu SC. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology 2013; 57:2299-313; PMID:23325601; http://dx.doi.org/10.1002/hep.26258
- Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T, Manabe T, et al. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J 2005; 24:963-73; PMID:15719014; http://dx.doi.org/10.1038/sj.emboj.7600588
- Sirirattanakul S, Wannakrairot P, Tencomnao T, Santiyanont R. Gene expression profile in breast cancer comprising predictive markers for metastatic risk. Genet Mol Res 2015; 14:10929-36; PMID:26400320; http://dx.doi.org/10.4238/2015.September.21.3
- Sangar F, Schreurs AS, Umaña-Diaz C, Clapéron A, Desbois-Mouthon C, Calmel C, Mauger O, Zaanan A, Miquel C, Fléjou JF, Praz. Involvement of small ArfGAP1 (SMAP1), a novel Arf6-specific GTPase-activating protein, in microsatellite instability oncogenesis. Oncogene 2014; 33:2758-67; PMID:23752192; http://dx.doi.org/10.1038/onc.2013.211
- Yagi R, Tanaka M, Sasaki K, Kamata R, Nakanishi Y, Kanai Y, Sakai R. ARAP3 inhibits peritoneal dissemination of scirrhous gastric carcinoma cells by regulating cell adhesion and invasion. Oncogene 2011; 30:1413-21; PMID:21076469; http://dx.doi.org/10.1038/onc.2010.522
- Junnila S, Kokkola A, Karjalainen-Lindsberg ML, Puolakkainen P, Monni O. Genome-wide gene copy number and expression analysis of primary gastric tumors and gastric cancer cell lines. BMC Cancer 2010; 10:73; PMID:20187983; http://dx.doi.org/10.1186/1471-2407-10-73
- Fan C, Tian Y, Miao Y, Lin X, Zhang X, Jiang G, Luan L, Wang E. ASAP3 expression in non-small cell lung cancer: association with cancer development and patients' clinical outcome. Tumour Biol 2014; 35:1489-94; PMID:24078447; http://dx.doi.org/10.1007/s13277-013-1205-1
- Chang J-S, Su C-Y, Yu W-H, Lee W-J, Liu Y-P, Lai T-C, Jan Y-H, Yang Y-F, Shen C-N, Shew J-Y, et al. GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget 2015; 6:36278-91; PMID:26462147
- Hou T, Yang C, Tong C, Zhang H, Xiao J, Li J. Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int J Clin Exp Pathol 2013; 7:280-7; PMID:24427349
- Willis S, Villalobos VM, Gevaert O, Abramovitz M, Williams C, Sikic BI, Leyland-Jones B, Siegel R, Naishadham D, Jemal A, et al. Single Gene Prognostic Biomarkers in Ovarian Cancer: A Meta-Analysis. PLoS One 2016; 11:e0149183; PMID:26886260; http://dx.doi.org/10.1371/journal.pone.0149183
- Ahn JY, Rong R, Kroll TG, Van Meir EG, Snyder SH, Ye K. PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. J Biol Chem 2004; 279:16441-51; PMID:14761976; http://dx.doi.org/10.1074/jbc.M312175200
- Ahn JY, Hu Y, Kroll TG, Allard P, Ye K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proc Natl Acad Sci USA 2004; 101:6993-8; PMID:15118108; http://dx.doi.org/10.1073/pnas.0400921101
- Knobbe CB, Trampe-Kieslich A, Reifenberger G. Genetic alteration and expression of the phosphoinositol-3-kinase/Akt pathway genes PIK3CA and PIKE in human glioblastomas. Neuro applied neurobiol 2005; 31:486-90; PMID:16150119; http://dx.doi.org/10.1111/j.1365-2990.2005.00660.x
- Xie Z, Jiang Y, Liao E-Y, Chen Y, Pennypacker SD, Peng J, Chang SM. PIKE mediates EGFR proliferative signaling in squamous cell carcinoma cells. Oncogene 2012; 31:5090-8; PMID:22349826; http://dx.doi.org/10.1038/onc.2012.10
- Cha JD, Kim HJ, Cha IH. Genetic alterations in oral squamous cell carcinoma progression detected by combining array-based comparative genomic hybridization and multiplex ligation-dependent probe amplification. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2011; 111:594-607; PMID:21334929; http://dx.doi.org/10.1016/j.tripleo.2010.11.020
- Sato H, Hatanaka KC, Hatanaka Y, Hatakeyama H, Hashimoto A, Matsuno Y, Fukuda S, Sabe H. High level expression of AMAP1 protein correlates with poor prognosis and survival after surgery of head and neck squamous cell carcinoma patients. Cell Commun Signal 2014; 12:17; PMID:24621372; http://dx.doi.org/10.1186/1478-811X-12-17
- Li M, Li M, Tian L, Tian L, Yao H, Yao H, Lu J, Lu J, Ge J, Ge J, et al. ASAP1 mediates the invasive phenotype of human laryngeal squamous cell carcinoma to affect survival prognosis. Oncol Rep 2014; 31:2676-82; PMID:24788532
- Hashimoto S, Mikami S, Sugino H, Yoshikawa A, Hashimoto A, Onodera Y, Furukawa S, Handa H, Oikawa T, Okada Y, et al. Lysophosphatidic acid activates Arf6 to promote the mesenchymal malignancy of renal cancer. Nat Commun 2016; 10656; PMID:26854204; http://dx.doi.org/10.1038/ncomms10656
- Lu X, Wan F, Zhang H, Shi G, Ye D. ITGA2B and ITGA8 are predictive of prognosis in clear cell renal cell carcinoma patients. Tumour Biol 2016; 37:253-62; PMID:26198048; http://dx.doi.org/10.1007/s13277-015-3792-5
- Cai Y, Wang J, Li R, Ayala G, Ittmann M, Liu M. GGAP2/PIKE-a directly activates both the Akt and nuclear factor-kappaB pathways and promotes prostate cancer progression. Cancer Res 2009; 69:819-27; PMID:19176382; http://dx.doi.org/10.1158/0008-5472.CAN-08-2537
- Nalla AK, Williams TF, Collins CP, Rae DT, Trobridge GD. Lentiviral vector-mediated insertional mutagenesis screen identifies genes that influence androgen independent prostate cancer progression and predict clinical outcome. Mol Carcinog 2015; PMID:26512949; http://dx.doi.org/10.1002/mc.22425.
- Lin D, Watahiki A, Bayani J, Zhang F, Liu L, Ling V, Sadar MD, English J, Fazli L, So A, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res 2008; 68:4352-9; PMID:18519696; http://dx.doi.org/10.1158/0008-5472.CAN-07-5237
- Huang C, Sheng Y, Jia J, Chen L. Identification of melanoma biomarkers based on network modules by integrating the human signaling network with microarrays. J Cancer Res Ther 2014; 10(Suppl):C114-24; PMID:25450268
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen I-M, Atlas SR, Kang H, Ar K, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 2010; 116:4874-84; PMID:20699438; http://dx.doi.org/10.1182/blood-2009-08-239681
- Yoo SM, Antonyak MA, Cerione RA. The adaptor protein and Arf GTPase-activating protein Cat-1/Git-1 is required for cellular transformation. J Biol Chem 2012; 287:31462-70; PMID:22807447; http://dx.doi.org/10.1074/jbc.M112.353615
- Okabe H, Furukawa Y, Kato T, Hasegawa S, Yamaoka Y, Nakamura Y. Isolation of development and differentiation enhancing factor-like 1 (DDEFL1) as a drug target for hepatocellular carcinomas. Int J Oncol 2004; 24:43-8; PMID:14654939
- Chen J, Yang P, Yang J, Wen Z, Zhang B, Zheng X. GIT1 is a novel prognostic biomarker and facilitates tumor progression via activating ERK/MMP9 signaling in hepatocellular carcinoma. Onco Targets Ther 2015; 8:3731-42; PMID:26719701
- Cai T, Xiao J, Wang ZF, Liu Q, Wu H, Qiu YZ. Identification of differentially coexpressed genes in gonadotrope tumors and normal pituitary using bioinformatics methods. Pathol Oncol Res 2014; 20:375-80; PMID:24198235; http://dx.doi.org/10.1007/s12253-013-9706-1
- Harada T, Chelala C, Crnogorac-Jurcevic T, Lemoine NR. Genome-Wide Analysis of Pancreatic Cancer Using Microarray-Based Techniques. Pancreatology 2009; 9:13-24; PMID:19077451; http://dx.doi.org/10.1159/000178871
