Abstract
Background
Progress in precision medicine relies on the access to, use of, and exchange of genomic and associated clinical data, including from children. The ethical, legal, and social issues (ELSI) of such data access, use, and exchange may be accentuated in the pediatric context due in part to the highly sensitive nature of genomic data, children’s consent-related vulnerabilities, and uncertain risks of reidentification. Systematic analyses of the ELSI and scientific reasons for why and how genomic data may be shared responsibly are, however, limited. Methods: We conducted a modified systematic review of reasons according to Sofaer and Strech to examine the ELSI and scientific reasons for “responsible” sharing of children’s genomic and associated clinical data. Empirical articles, commentaries, and data-sharing policies indexed in Medline, Scopus, Web of Science, and BIOSIS were included in the analysis if they discussed ELSI and were published between 2003 and 2017 in English. Results: One hundred and fifty-one records met our inclusion criteria. We identified 11 unique reasons and 8 subreasons for why children’s genomic data should or should not be shared. Enhancing the prospect of direct and indirect benefits and maximizing the utility of children’s data were top reasons why data should be shared. Inadequate data privacy protection was the leading reason why it should not. We furthermore identified 8 reasons and 30 subreasons that support conditional data sharing, in which recontact for the continued use of children’s data once they reach the age of majority was the most frequently endorsed condition. Conclusions: The complete list of ELSI reasons and responsible conditions provides an evidentiary basis upon which institutions can develop data-sharing policies. Institutions should encourage the sharing of children’s data to advance genomic research, while heeding special reconsent and data protection mechanisms that may help mitigate uncertain longitudinal risks for children and families.
Introduction
Translating discoveries in precision medicine to achieve improved individual- and population-level health outcomes necessitates broad sharing of genomic and associated clinical data. Such sharing is especially pertinent during childhood, when many Mendelian and other heritable genetic conditions first present and may be clinically actionable. Indeed, advancements in understanding genomic etiologies of childhood diseases as a result of concerted data sharing are innumerable (Downing et al. Citation2012; Mefford, Batshaw, and Hoffman Citation2012; Beaulieu et al. Citation2014), and whole genome sequencing used as a first-in-line test has been shown to increase diagnostic yield (Schofield et al. Citation2010; Xue et al. Citation2015), sometimes by nearly 27% (Lindstrand et al. Citation2019). The clinical successes of sequencing have furthermore motivated Congressional action (H.R.4144 116th Congress Citation2019). For children suspected of suffering from rare, undiagnosed genetic conditions, in particular, sharing their linked genotypic and phenotypic data in accessible variant databases may afford them the only opportunity for accurate diagnosis (Beaulieu et al. Citation2014; Boycott et al. Citation2014; ACMG Board of Directors Citation2017; Might and Wilsey Citation2014). Genomic data are, however, inherently identifying, and their broad exchange can pose informational risks not only to the pediatric patients, but also to their biological relatives (McGuire, Oliver, et al. Citation2011; Kaye Citation2012).
These general ethical, legal, and social issues (ELSI) give rise to unique considerations when data sharing involves children. Children’s inability to legally consent underpins their status as a vulnerable population in data-intensive research. While parents are authorized to make decisions in the best interests of their child, it is generally accepted that children assume decision-making rights once they reach the age of majority. Practical challenges can limit data-sharing preferences of future adults (Knoppers et al. Citation2016)—for example, when data are contributed anonymously and then aggregated for inclusion in larger, combined datasets. This aggregation makes retrieval and removal therefore unrealistic. Recent reports of patient reidentification from datasets originally thought to be anonymized further heighten data security concerns (Gymrek et al. Citation2013; Erlich et al. Citation2018), and the accuracy with which patients can be reidentified from such datasets forecloses on what scholars term the child’s “right to an open future” (Davis Citation2009; Davis Citation1997). Parents face ethical tensions, as enhancing immediate clinical benefits, such as diagnosis, may involve putting their child’s data at uncertain risk with little longitudinal control over how the data will be exchanged in the future. It is primarily this specific circumstance and those related to sharing genomic data from banked biospecimens involving children that this systematic review of the literature considers in depth.
Despite the need for practical guidance on how to responsibly share children’s genomic data, few analyses have sought to landscape ELSI reasons for responsible sharing as expressed in both the empirical bioethics and gray literature. Absent such analysis, institutions may inappropriately restrict or aimlessly permit access to sensitive genomic data. Both outcomes have negative consequences on advancing knowledge in pediatric disease, and ultimately misguide evidence-based policy development for responsible data sharing in the data-intensive sciences such as genomics.
Review methods/search
We conducted a systematic review of reasons according to Sofaer and Strech (Citation2011) to fill the aforementioned knowledge gap and to guide institutional best practices for sharing genomic data involving children. Systematic reviews of reasons “take into account the specific conceptual and practical challenges of empirical bioethics” (Strech and Sofaer Citation2012), while preserving the systematicity associated with traditional reviews. This method of review enables reviewers to systematically search for and contextualize evidence contained in argument-based literatures that can be used to prioritize agenda setting and motivate policy action on complex issues that span medical, legal, and philosophical disciplines. Systematic reviews of reasons make arguments the primary outcomes of interest. The Sofaer and Strech model proposes four steps:
Formulate the review question and eligibility criteria.
Identify all of the literature that meets the eligibility criteria.
Extract and synthesize data.
Derive and present results: the answer to the review question.
Our synthesis of the data in step 3 involved first identifying passages where a reason is expressed (called a reason mention). We subsequently categorized reason mentions into two “types” (either a reason, or a subreason)Footnote1. While Sofaer and Strech do not prescribe any one qualitative approach to categorizing reason types, all approaches aim to “compare text passages that mention reasons across papers and to match reason mentions from one paper with reason mentions from another, ensuring that a reason type captures similar reason mentions from different papers” (Strech and Sofaer Citation2012, 123).
Our review departs from the Sofaer and Strech (Citation2011) model in that we ask two questions of the literature to more comprehensively capture the ELSI of “responsible” genomic data sharing involving children. We supplemented a traditional, reason-based review question—Which reasons have been given to support the view that children’s genomic and associated clinical data should/not be shared?—with a secondary review question that asked, What conditions are needed to support responsible sharing of children’s genomic and associated clinical data? Early data analyses informed this decision, as they revealed greater contention around the conditions that support responsible data sharing, rather than whether data should be shared ipso facto. Put simply, our results suggest broad support for sharing genomic data but is contingent on certain safeguards. The second review question details those conditions as reflected in the literature, and documents their supporting rationales. It is important to note a conceptual difference between conditions that support “responsible” and compliant data sharing. The latter conditions are both jurisdiction specific and institution specific. Compliant data sharing appeals to regulations governing human subjects research and data protection, among others, and for this reason is outside the authority of this article to define.
With support from a reference librarian, the review team searched the empirical and gray literatures indexed in MedLine, BIOSIS, Scopus, and Web of Science for records published in English between 2002 and 2017. A detailed search strategy for each database is provided in the supplementary materials. We included empirical studies; commentaries; editorials or position statements; conference proceedings; professional organization reports; and international or professional guidelines if they
Discussed ethical, legal, social implications (ELSI) of exchanging whole genome/exome data involving children.
The sharing of genomic or associated clinical data involved patients aged 0–18 years old (neonates to the age of majority in most jurisdictions).
The sharing of genomic or associated clinical data was derived in the context of either pediatric clinical research or pediatric care.
Records were excluded if they discussed pediatric data sharing only in relation to the return of incidental findings/individual research results or in newborn screening. Previously published systematic reviews (Mackley et al. Citation2017; Bertier et al. Citation2017) and empirical studies (McGowan et al. Citation2018; Burke et al. Citation2013; Holm Citation2017; Bishop, Strong, and Dimmock Citation2017; Petersen et al. Citation2017; Driessnack and Gallo Citation2011) address these issues at length. We also excluded technical papers where interoperability or security measures were detailed without explicit discussion of ELSI motivating these data protective measures.Footnote2
Data analysis
Lead author VR conducted the analysis of subreason/reason mentions on all retained records and policy documents using the NVivo 11 software. VR first scanned the document for all mentions of “data shar*,” “pediatric,” “ethic*,” “legal,” or “social.” Using qualitative content analysis (Sandelowski Citation2000), relevant reasons were coded to produce a preliminary coding scheme. All members of the review team consulted on the final codebook that was subsequently used to analyze subreason/reason mentions in the entire dataset.
Systematic review results
Publication characteristics
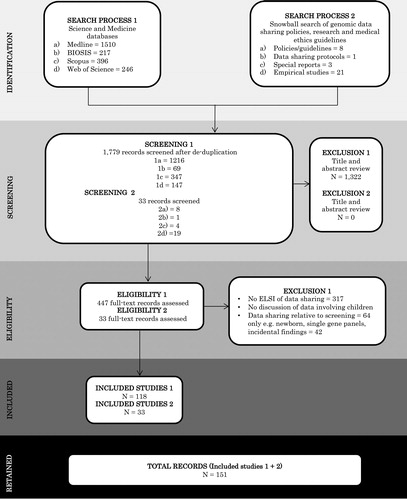
In total, 151 records met our inclusion criteria; 118 records were retained from the database search, the bibliographies, and in-text references, which authors VR and GB hand searched for any normative guidance documents, such as policies, guidelines, or conventions, cited in the record. This snowball search resulted in an additional 33 records. provides a PRISMA summary of the search results, and a data extraction table of the following characteristics can be found in :
Table 1. Publication characteristics
Publication type.
Content type.
Journal field.
Context in which pediatric data sharing was primarily discussed.
Sixty-five (43%) records discussed data sharing in an ELSI commentary/opinion piece, while 27 (18%) discussed ELSI in the context of organized data-sharing programs or pediatric research consortia. All 43 records reporting empirical data were critically appraised using the Mixed Methods Appraisal Tool (MMAT). Of these, 39 (94%) earned a MMAT score of 0.75 or higher (see Supplementary Material). Four records presented results from two empirical studies (Burstein et al. Citation2014; McGuire, Oliver, et al. Citation2011; Hens et al. Citation2009a; Hens et al. Citation2009b). Fourteen percent of all records for which a publication year was available were published in 2016; nearly half (49%) were published in between 2013 and 2017, inclusively.
Subreason/reason identity and incidence
We identified 604 total subreason/reason mentions, which we grouped under 19 main reason types and 38 subreason types. Enhance the prospect of direct and indirect benefits to individual as well as other pediatric patients was the reason most mentioned (27 mentions) for why children’s data should be shared. Consent was the condition most mentioned (29 mentions) supporting responsible sharing. The right to recontact/reconsent to continued data use at the age of majority garnered the highest number of subreason mentions (35) related to conditions that support responsible sharing, followed by limiting parental authority for broad consent (29) and managing data access via user restrictions (29).
Findings
Which reasons have been given to support the view that children’s genomic and associated clinical data should/not be shared?
Of 151 records that met our inclusion criteria, we identified one reason mention for why children’s data should categorically not be shared: “If whole genome sequencing to identify preventable diseases or SNPs or haplotypes associated with drug responsiveness does occur, the child’s DNA should be discarded afterwards and genomic information erased or stringently protected. If protective measures are not feasible, a decision will have to be made whether to genotype them at all” (Robertson Citation2003, 7).Two subreasons underpin this position: a lack of a compelling need to retain results derived from genotyping a patient who at present lacks competency to consent, and the potential for data misuse. The author reasons that whole genome sequencing for those patients rendered incompetent to consent at the time of genotyping should be reserved for rare circumstances when immediate clinical benefit justifies sharing, or when genome-wide comparisons are the primary aim of the research. Narrower genetic tests with time-limited information storage could suffice for meeting the direct clinical needs for the child, the author reasons, and therefore such options should be exhausted before considering broad sharing.
Time and regulatory history are key factors when interpreting the preceding subreasons/reasons. The year of publication (2003) marked the completion of the Human Genome Project, and the applications of whole genome sequencing in the clinic elicited both awe and trepidation. This record also predated important privacy legislations—most importantly the Genetic Information Nondiscrimination Act (GINA)—and human subjects research protections in the Revised Common Rule. Both GINA and the Revised Common Rule later included specific protections for genetic/genomic data, including for children. We posit that a stronger regulatory climate might have instilled greater trust in the data protections afforded under the law, and in turn a more permissive stance toward data sharing with appropriate oversight.
We identified 10 reasons and 7 subreasons why pediatric genomic and associated clinical data should be shared (), with a total of 149 mentions. Enhancing the prospect of benefit to individual and future pediatric patients was the reason most frequently cited for why pediatric data should be shared (27 reason mentions), followed by the need to maximize the scientific and clinical utility of children’s data (13 reason mentions).
Table 2. Reasons and subreasons given for why children’s genomic and associated clinical data should (n = 3) and should not (n = 149) be shared [number of mentions].
Table 3. Reasons and subreasons given for the conditions that support “responsible” sharing of children’s genomic and associated clinical dataFootnote3 [number of mentions] (Sofaer and Strech Citation2011).
The interests of the child were often raised conjointly with the types and likelihoods of direct benefits anticipated, particularly when data are collected in clinical contexts. We subdivided the mention, Data sharing benefits, to distinguish among three discrete types of benefits. These include direct (7 subreason mentions), indirect (12 subreason mentions), and both indirect and direct benefits (8 subreason mentions). We also distinguished between intended beneficiaries of those benefits, including individual patients and other/future children. Twelve reason mentions endorsed data sharing on the basis that it may generate indirect benefits to other and future children, including one subreason that sharing manifests the solidarity principle.
Of 12 reason mentions that supported data sharing on the basis that it advances pediatric medicine, 4 subreasons explicitly discussed improving diagnostic yields and furthering pediatric-specific drug development (5 subreason mentions). While data protection and uncertain informational risks were previously cited as reasons against sharing, four reason mentions concluded that informational risks are reduced when sharing data that are “special or that would be difficult or expensive to duplicate” (Birmingham and Doyle Citation2009, 46). Achieving necessary sample sizes considering the relative rarity of some pediatric conditions substantiated 13 reason mentions given for why scientific collaboration vis-à-vis data sharing was both a scientific and ethical need in practice (Mccormack et al. Citation2013; Anonymous Citation2016).
What conditions support “responsible” sharing of children’s genomic and associated clinical data?
We categorized 442 total reason mentions into 8 main reasons, and 25 subreasons for why genomic data sharing should be conditional. organizes these subreasons/reasons by frequency of mentions. The most-mentioned subreason/reason identified in our analysis centered on conditions related to consent. We identified 95 total subreason/reason mentions related to data protection, 69 related to informational risk evaluation, 47 concerning genomic data management, and 39 for data access. The involvement of community stakeholder groups in establishing responsible data-sharing practices comprised 3 additional reasons, namely, the importance of community outreach and continued education (13), stakeholder willingness and trust (17), and incentivizing researchers to share via appropriate attribution (6).
Consent models
Debate on whether and for how long parents should authorize continued use of pediatric data at the age of majority (29 subreason mentions) was among the most mentioned practices supporting conditional data sharing; the most heterogeneous consent modality related to broad consent (11 subreason mentions). There was general consensus that broad consent referred to a one-time permission to reuse data for future research, pending requisite ethics oversight (Riggs et al. Citation2019; Sanderson et al. Citation2017). Importantly, eight reason mentions discussed various waivers of consent to facilitate greater access to larger datasets while still satisfying permissions outlined in original consent documents. Irrespective of consent model, 27 subreasons supported that children should be involved in decision-making processes in developmentally appropriate ways, and 35 strongly endorsed recontacting children at the age of majority when logistically possible.
Responsible data protection
Nine reason mentions claimed that genetic/genomic data categorically warrants special protections above those required for other health-related data due to its inherent identifiability. This position was irrespective of whether the data contributor was considered part of a vulnerable population. Generally, data protections were considered proportionate to the (i) sensitivities of data shared, (ii) intended purposes for sharing, and (iii) likelihood of unauthorized reidentification (Brothers et al. Citation2014; Fisher, McCarthy, and Harrington Citation2013; Hens, Lévesque, et al. Citation2011; Scholtens et al. Citation2015; SACHRP Citation2018; Brothers Citation2011). Records argued that data protection responsibilities rest with clinical professionals (World Medical Association Citation2013) or data stewards in research contexts (Manolio et al. Citation2007). It is also worth noting that privacy and data protection were often discussed interchangeably, despite their conceptual differences (Gostin Citation1995).
The most mentioned reason (25) involved data deidentification, under which 10 subreason mentions justified how technical deidentification strategies satisfy both regulatory requirements and overall data utility. Although linking multiple datasets together was perceived to enhance the likelihood of reidentifiability (19 subreason mentions), such linkage was overwhelmingly endorsed in combination with adequate data governance and oversight (Canadian Institutes of Health Research Natural Sciences and Engineering Research Council of Canada and Social Sciences and Humanities Research Council of Canada Citation2014; Lea Citation2013; Fisher, McCarthy, and Harrington Citation2013; Lloyd et al. Citation2015; Platt et al. Citation2014; Driessnack and Gallo Citation2011; Giesbertz, Bredenoord, and van Delden Citation2015; Heeney et al. Citation2011; Suresh et al. Citation2005; Mascalzoni, Paradiso, and Hansson Citation2014; Majumder et al. Citation2017; Ebner et al. Citation2016; Wjst Citation2010; Said et al. Citation2017; Manolio et al. Citation2007; Brett and Deary Citation2014). Our finding is consistent with ongoing initiatives to improve technical infrastructures (Wan et al. Citation2017; Decouchant et al. Citation2018; Humbert et al. Citation2017).
Benefits and risks
We found three distinct types of risk–benefit balances: (1) protecting children and advancing the science of pediatric disease (Bertier et al. Citation2017; Manhas et al. Citation2016; McGuire, Oliver, et al. Citation2011; Burstein et al. Citation2014; Nature Biotechnology Citation2015; Hens, Lévesque, et al. Citation2011; Platt et al. Citation2017; Fisher, McCarthy, and Harrington Citation2013; Hens, Wright, and Dierickx Citation2009; Dowty and Korff Citation2009; Manolio et al. Citation2007); (2) respecting individual rights versus serving public interests (Ahrens et al. Citation2006; Petrini et al. Citation2012); and (3) preserving data privacy versus activating a right to data access (Golding Citation2009; Nooner et al. Citation2012). The majority of records implied that balancing benefits with informational risks (17 reason mentions) is complicated, given the uncertainties of data privacy/and security at the time of data contribution (Dowty and Korff Citation2009; Bertier et al. Citation2017; Manhas et al. Citation2016; McGuire, Oliver, et al. Citation2011; Platt et al. Citation2014; Burstein et al. Citation2014; Fisher, McCarthy, and Harrington Citation2013; Hens, Wright, and Dierickx Citation2009; Hens, Lévesque, et al. Citation2011; Nature Biotechnology Citation2015; Nooner et al. Citation2012; Ahrens et al. Citation2006; Anderson and Merry Citation2009; Golding Citation2009; Lunshof and Gurwitz Citation2013; Manolio et al. Citation2007; Petrini et al. Citation2012; Hens, Lévesque, et al. Citation2011). Threats to loss of privacy were discussed in general terms as reasons on which to condition data sharing (12 mentions), whereas unauthorized data access by third parties was the most mentioned subreason for this loss (15). Appealing to the argument that benefit–risk evaluation should be based on current available evidence, 12 subreason mentions indicated that sharing genomic data was, on the whole, considered minimal risk, especially when data were deidentified (25 subreason mentions).
Responsible data management
The records we synthesized interpreted data management to include administrative tasks that support data formatting and standardization for exchange of data across information systems (9 reason mentions). We identified two management approaches to achieve this level of data management: via mechanisms of mutual accountability for both data sharers and users (3 subreason mentions), and by improving data interoperability and ease of use (13 subreason mentions). Four subreason mentions explicitly endorsed a shared lexicon to facilitate such interoperability (see, e.g., the Global Alliance for Genomics and Health (GA4GH)) Citation2016). Clarifying data ownership was an additional responsibility of data managers, according to 7 subreason mentions. Several records outlined conflicting viewpoints on whether patients or institutions own the shared data. Authors supporting the former argued that data derived from biospecimens are an extension of the patient’s physical body and patients retain rights to control the data produced. Others reasoned that entities that expend resources to maintain samples/data ultimately own them, as well as all commercial/financial benefits derived therefrom.
Responsible data access
Conditions for responsible access were contingent on the data type, where the data were being deposited, and by whom. Controlled access via user restrictions was the most mentioned subreason (29) supporting responsible access to identifiable or coded data. Many records discussed user authentication and verification (Dyke et al. Citation2016) as practical means by which to ensure bona fide users—for example, researchers, physicians, and so on can access data of a potentially sensitive nature. Seven of 10 reason mentions described the role of data access committees, data use agreements, and other confidentiality agreements to manage controlled access requests (Pediatric Imaging Citation2011; Ries, LeGrandeur, and Caulfield Citation2010; Birmingham and Doyle Citation2009; Wjst Citation2010; Fisher, Bromley, and Mansfield Citation2016; Jernigan et al. Citation2016; Manolio et al. Citation2007), while 3 subreason mentions named ethics review boards as primary gatekeepers of data access (Pinto et al. Citation2015; Mccabe and Mccabe Citation2011; Tindana et al. Citation2012).
Stakeholder involvement
Of 10 reason mentions centered on sustaining patient willingness and trust in the research and clinical enterprise, 7 subreason mentions proposed specific methods for improving patient engagement. Direct stakeholder involvement, either as part of the shared decision-making process, or as participants in future public perceptions research, was among the most frequently mentioned methods. These methods were separate from continuing education and stakeholder outreach (13 reason mentions), including educating patients about their rights to access their own medical records that might contain genomic data, and more informed ELSI guidance through public priority setting in research. Unsurprisingly, researchers were among the most frequently discussed stakeholder communities among which to foster responsible data sharing practices. Six reason mentions advocated for improved data attribution as an incentive structure for researchers to share raw genomic data, especially in the context of large national genomics initiatives (Holland et al. Citation2009; Majumder et al. Citation2017), pursuant to Article 27(General Assembly of the United Nations Citation1948) of the Universal Declaration of Human Rights. Information regarding data release and the associated structures governing such release (10 reason mentions) both played central roles in enhancing patient willingness and public trust.
Limitations
Several limitations should be noted when interpreting our results. First, our review questions demanded inclusion criteria that were broad enough to capture relevant ELSI reasons across an interdisciplinary array of literatures. For similar reasons, we did not distinguish between types of data-sharing environments in our analysis, such as research only or clinic only. Such distinctions certainly impact the generalizability of our findings to other pediatric patient populations, legal jurisdictions, and data use cases. Two reviewers resolved discrepancies in the qualitative coding prior to analysis, but it is possible that different reviewers would make different reason assignments. That is, “[reason] types could be made narrower or broader; some broad types cover diverse narrow types; and there may be good reasons to class a narrow type of reason under different broad types” (Sofaer and Strech Citation2011, 178). As a result, the sum totals for reasons reported in this article would also change. Lastly, the conditions we described that support responsible sharing suggest there are many negative consequences of sharing for children and their families if such conditions are not met. We do not consider these consequences, in and of themselves, reasons against data sharing. Rather higher frequency with which some reason/subreason mentions appear in the literature may suggest, for instance, how the data-sharing community weighs the significance of those conditions or reflect the most publicized reasons for conditional sharing.
Conclusion
This review is an evolving synthesis of ELSI reasons that ground “responsible” data sharing in theory and practice. It not only identifies specific practices that support responsible sharing but also contributes to the policy conversation by highlighting ELSI priorities around which to base empirical research for policymaking purposes. We thematically categorized 58 unique subreasons/reasons for why, and under what conditions, to support responsible data sharing into the following groups: (1) parent and family involvement, (2) stakeholder involvement, (3) benefits and risks and (4) data governance and release (). Our conclusions from this review later informed an ethical–legal framework of responsible data sharing for pediatric genomics published elsewhere (Rahimzadeh et al. Citation2018).
Maximizing clinical and scientific utility of children’s data was the most mentioned reason for why children’s data should be shared. The convergence of the evidence-based medicine movement and rapid advances in computational power since first sequencing the human genome in 2003 could have underscored why the aforementioned reason was most frequently discussed. Inadequate data protection was the foremost reason cited for why data should not be shared, as well as a primary theme supporting reasons for conditional sharing. Ensuring that consent reflects the potential for future secondary use via recontact at the age of majority was the most mentioned subreason for conditional data sharing.
Taken together, the complete list of reasons and conditions this review synthesizes makes two practical contributions to the field. First, it guides pediatric research institutions in developing data-sharing policies that align with contemporary ELSI priorities and scientific aims (see, e.g., Taylor et al. Citation2019). Second, our review supports current and future policy decision makers by identifying data-sharing practices that, based in part on the frequency of reasons identified, could be strengthened. Recontact at the age of majority and data access restrictions are two noteworthy examples in this regard. Future empirical research is needed involving individuals whose genomic data were shared as children to assess how the ELSI reasons identified herein translate across the life course.
Author contributions
Authors VR, GB, and BMK conceptualized the literature review study as part of a doctoral dissertation project. VR and GB screened articles for inclusion and developed the coding strategy for the content analysis of all retained articles. VR led in the drafting of the article and peer review revisions. All authors approved the final version of the article.
Ethical approval
This study was approved by the McGill University Institutional Review Board.
uabr_a_1818875_sm2090.zip
Download Zip (132.4 KB)Acknowledgments
The authors sincerely thank the anonymous peer reviewers for their insightful comments and rigorous review of earlier versions of this article, as well Genevieve Gore, chief librarian for the McGill University Department of Family Medicine, for her guidance in developing the search strategy.
Conflicts of interest
No potential conflict of interest was reported by the authors.
Additional information
Funding
Notes
1 Sofaer and Strech provide examples both broad and narrow reason “types.” We distinguish between these different types as “reasons” and “subreasons,” respectively, in the review. A reason (e.g., avoid exploitation) is thematically related to, but conceptually broader than, a subreason (e.g., avoid exploiting research participants).
2 While return of incidental or secondary findings is considered a form of data sharing, this review of reasons focused on responsible sharing outside the nucleus of care. Similarly, newborn screening programs may imply responsible sharing, yet are obligatory forms of data collection and management mandated by the state and under the auspices of public health.
3 The total number of reason mentions is not the sum total of its subreason mentions for many of the conditions analyzed here. This is because a main reason was counted if the record discussed the condition but did not specify how to enable this condition. For example, we identified 10 reason mentions for data access as a feature of responsible sharing; 29 subreason mentions explicitly suggested user restrictions to facilitate such data access. We assigned the latter (data user restriction) as a subreason under the main reason category responsible data access.
References
- ACMG Board of Directors. 2017. Laboratory and clinical genomic data sharing is crucial to improving genetic health care: A position statement of the American College of Medical Genetics and Genomics. Genetics in Medicine 19:721–727. doi: 10.1038/gim.2016.196.
- Ahrens, W., K. Bammann, S. de Henauw, J. Halford, A. Palou, I. Pigeot, A. Siani, and M. Sjöström. 2006. Understanding and preventing childhood obesity and related disorders-IDEFICS: A European multilevel epidemiological approach. Nutrition, Metabolism, and Cardiovascular Diseases: NMCD 16 (4):302–8. doi: 10.1016/j.numecd.2006.01.011.
- Anderson, B. J., and A. F. Merry. 2009. Data sharing for pharmacokinetic studies. Paediatric Anaesthesia 19 (10):1005–10. doi: 10.1111/j.1460-9592.2009.03051.x.
- Anonymous. 2016. Data sharing pooling clinical details helps doctors to diagnose rare diseases: But More Sharing Is Needed. Nature 534:5–6.
- Beaulieu, C. L., J. Majewski, J. Schwartzentruber, M. E. Samuels, B. A. Fernandez, F. P. Bernier, M. Brudno, B. Knoppers, J. Marcadier, D. Dyment, et al. 2014. FORGE Canada consortium: Outcomes of a 2-year national rare-disease gene-discovery project. American Journal of Human Genetics 94 (6):809–17. doi: 10.1016/j.ajhg.2014.05.003.
- Bertier, G., K. Sénécal, P. Borry, and D. F. Vears. 2017. Unsolved challenges in pediatric whole-exome sequencing: A literature analysis. Critical Reviews in Clinical Laboratory Sciences 54 (2):134–9. doi: 10.1080/10408363.2016.1275516.
- Birmingham, K., and A. Doyle. 2009. Ethics and governance of a longitudinal birth cohort. Paediatric and Perinatal Epidemiology 23 (Suppl. 1):39–50. doi: 10.1111/j.1365-3016.2008.00995.x.
- Bishop, C. L., K. A. Strong, and D. P. Dimmock. 2017. Choices of incidental findings of individuals undergoing genome wide sequencing, a single center's experience. Clinical Genetics 91 (1):137–40. doi: 10.1111/cge.12829.
- Boycott, K. M., D. A. Dyment, S. L. Sawyer, M. R. Vanstone, and C. L. Beaulieu. 2014. Identification of genes for childhood heritable diseases. Annual Review of Medicine 65:19–31. doi: 10.1146/annurev-med-101712-122108.
- Brett, C. E., and I. J. Deary. 2014. Realising health data linkage from a researcher’s perspective: Following up the 6-day sample of the Scottish Mental Survey 1947. Longitudinal and Life Course Studies 5 (3):283–98. doi: 10.14301/llcs.v5i3.266.
- Brothers, K. B., J. A. Lynch, S. A. Aufox, J. J. Connolly, B. D. Gelb, I. A. Holm, S. C. Sanderson, J. B. McCormick, J. L. Williams, W. A. Wolf, et al. 2014. Practical guidance on informed consent for pediatric participants in a biorepository. Mayo Clinic Proceedings 89 (11):1471–80. doi:10.1016/j.mayocp.2014.07.006.Practical.
- Brothers, K. B. 2011. Biobanking in pediatrics: The human nonsubjects approach. Personalized Medicine 8 (1):79. doi: 10.2217/pme.10.70.
- Burke, W., A. H. M. Antommaria, R. Bennett, J. Botkin, E. W. Clayton, G. E. Henderson, I. A. Holm, G. P. Jarvik, M. J. Khoury, B. M. Knoppers, et al. 2013. Recommendations for returning genomic incidental findings? We need to talk! Genetics in Medicine 15 (11):854–9. doi: 10.1038/gim.2013.113.
- Burstein, M. D., J. O. Robinson, S. G. Hilsenbeck, A. L. McGuire, and C. C. Lau. 2014. Pediatric data sharing in genomic research: Attitudes and preferences of parents. Pediatrics 133 (4):690–7. doi: 10.1542/peds.2013-1592.
- Canadian Institutes of Health Research Natural Sciences and Engineering Research Council of Canada and Social Sciences and Humanities Research Council of Canada. 2014. The Tri council policy statement: Ethical conduct for research involving humans. https://ethics.gc.ca/eng/documents/TCPS_2-2014_FINAL_Web.pdf.
- Davis, D. S. 1997. Genetic dilemmas and the child’s right to an open future. The Hastings Center Report 27 (2):7–15. doi: 10.2307/3527620.
- Davis, D. S. 2009. Parental investment factor and the child’s right to an open future. Hastings Center Report 39 (2):24–7.
- Decouchant, J., M. Fernandes, M. Völp, F. M. Couto, and P. Esteves-Veríssimo. 2018. Accurate filtering of privacy-sensitive information in raw genomic data. Journal of Biomedical Informatics 82:1–12. doi: 10.1016/j.jbi.2018.04.006.
- Downing, J. R., R. K. Wilson, J. Zhang, E. R. Mardis, C.-H. Pui, L. Ding, T. J. Ley, and W. E. Evans. 2012. The pediatric cancer genome project. Nature Genetics 44 (6):619–22. doi: 10.1038/ng.2287.
- Dowty, T., and D. Korff. 2009. Protecting the virtual child: The law and children’s consent to sharing personal data. http://medconfidential.org/wp-content/uploads/2013/03/Protecting-the-virtual-child.pdf.
- Driessnack, M., and A. M. Gallo. 2011. Stop, look, and listen: Revisiting the involvement of children and adolescents in genomic research. Annual Review of Nursing Research 29:133–49. doi: 10.1891/0739-6686.29.133.
- Dyke, Stephanie, O. M., Dove, E. S., and B. M. Knoppers. 2016. Sharing health-related data: A privacy test? Genome Medicine 1:16024. doi: 10.1038/npjgenmed.2016.24.
- Ebner, H., D. Hayn, M. Falgenhauer, M. Nitzlnader, G. Schleiermacher, R. Haupt, and G. Erminio. 2016. Piloting the European unified patient identity management (EUPID) concept to facilitate secondary use of neuroblastoma data from clinical trials and biobanking. In 10th Annual Conference on Health Informatics Meets EHealth, EHealth 2016, eds., E. Ammenwerth, G. Schreier, A. Horbst, and D. Hayn. Vienna, Austria: Schönbrunn Palace. doi: 10.3233/978-1-61499-645-3-31.
- Erlich, Y., T. Shor, I. Pe’er, and S. Carmi. 2018. Identity inference of genomic data using long-range familial searches. Science (New York, NY) 362 (6415):690–4. doi: 10.1126/science.aau4832.
- Fisher, C. B., E. L. H. McCarthy, and E. L. Harrington. 2013. Ethics in prevention science involving genetic testing. Prevention Science: The Official Journal of the Society for Prevention Research 14 (3):310–8. doi: 10.1007/s11121-012-0318-x.
- Fisher, J. K., R. L. Bromley, and B. C. Mansfield. 2016. My retina TrackerTM: An on-line international registry for people affected with inherited orphan retinal degenerative diseases and their genetic relatives: A new resource. In Retinal degenerative disease, advances in experimental medicine and biology, ed. C. Bowes Rickman, 245–51. Cham: Springer International Publishing Switzerland.
- General Assembly of the United Nations. 1948. Universal declaration of human rights. Washington, DC: United Nations.
- Giesbertz, N. A. A., A. L. Bredenoord, and J. J. van Delden. 2015. Consent procedures in pediatric biobanks. European Journal of Human Genetics: EJHG 23 (9):1129–34. doi: 10.1038/ejhg.2014.267.
- Global Alliance for Genomics and Health (GA4GH). 2016. Data sharing lexicon. https://www.ga4gh.org/wp-content/uploads/GA4GH_Data_Sharing_Lexicon_Mar15.pdf.
- Golding, J. 2009. Data organisation and preparation for statistical analysis in a longitudinal birth cohort. Paediatric and Perinatal Epidemiology 23 (Suppl 1):219–25. doi: 10.1111/j.1365-3016.2009.01019.x.
- Gostin, L. O. 1995. Genetic privacy. The Journal of Law, Medicine & Ethics: A Journal of the American Society of Law, Medicine & Ethics 23 (4):320–30. doi: 10.1111/j.1748-720X.1995.tb01374.x.
- Gymrek, M., A. L. McGuire, D. Golan, E. Halperin, and Y. Erlich. 2013. Identifying personal genomes by surname inference. Science (New York, NY) 339 (6117):321–4. doi: 10.1126/science.1229566.
- H.R.4144 116th Congress. 2019. Ending the Diagnostic Odyssey Act of 2019. https://www.congress.gov/bill/116th-congress/house-bill/4144/text?format=txt.
- Heeney, C., N. Hawkins, J. de Vries, P. Boddington, and J. Kaye. 2011. Assessing the privacy risks of data sharing in genomics. Public Health Genomics 14 (1):17–25. doi: 10.1159/000294150.
- Hens, K., H. Nys, J.-J. Cassiman, and K. Dierickx. 2009a. Genetic research on stored tissue samples from minors: A systematic review of the ethical literature. American Journal of Medical Genetics. Part A 149A (10):2346–58. doi: 10.1002/ajmg.a.33032.
- Hens, K., H. Nys, J.-J. Cassiman, and K. Dierickx. 2009b. Biological sample collections from minors for genetic research: A systematic review of guidelines and position papers. European Journal of Human Genetics: EJHG 17 (8):979–90. doi: 10.1038/ejhg.2009.9.
- Hens, K., E. Lévesque, and K. Dierickx. 2011. Children and biobanks: A review of the ethical and legal discussion. Human Genetics 130 (3):403–13. doi: 10.1007/s00439-011-1031-8.
- Hens, K., H. Nys, J. J. Cassiman, and K. Dierickx. 2011. Risks, benefits, solidarity: A framework for the participation of children in genetic biobank research. The Journal of Pediatrics 158 (5):842–8. doi: 10.1016/j.jpeds.2010.12.036.
- Hens, K., J. Wright, and K. Dierickx. 2009. Biobanks: Oversight offers protection. Science (New York, NY) 326 (5954):798–9. doi: 10.1126/science.326_798c.
- Holland, A., J. Whittington, O. Cohen, L. Curfs, F. Delahaye, O. Dudley, B. Horsthemke, A.-C. Lindgren, C. Nourissier, N. Sharma, et al. 2009. The European Prader-Willi Syndrome clinical research database: An aid in the investigation of a rare genetically determined neurodevelopmental disorder. Journal of Intellectual Disability Research: JIDR 53 (6):538–47. doi: 10.1111/j.1365-2788.2009.01172.x.
- Holm, I. A. 2017. Pediatric issues in return of results and incidental findings: Weighing autonomy and best interests. Genetic Testing and Molecular Biomarkers 21 (3):155–8. doi: 10.1089/gtmb.2016.0414.
- Humbert, M., E. Ayday, J.-P. Hubaux, and A. Telenti. 2017. Quantifying interdependent risks in genomic privacy. ACM Transactions on Privacy and Security 20 (1):1–31. doi: 10.1145/3035538.
- Jernigan, T. L., T. T. Brown, D. J. Hagler, N. Akshoomoff, H. Bartsch, E. Newman, W. K. Thompson, C. S. Bloss, S. S. Murray, N. Schork, et al. 2016. The pediatric imaging, neurocognition, and genetics (PING) data repository. NeuroImage 124 (Pt B):1149–54. doi: 10.1016/j.neuroimage.2015.04.057.
- Kaye, J. 2012. The tension between data sharing and the protection of privacy in genomics research. Annual Review of Genomics and Human Genetics 13:415–31. doi: 10.1146/annurev-genom-082410-101454.
- Knoppers, B. M., K. Sénécal, J. Boisjoli, P. Borry, M. C. Cornel, C. V. Fernandez, J. Grewal, I. A. Holm, E. Nelson, W. Pinxten, et al. 2016. Recontacting pediatric research participants for consent when they reach the age of majority. IRB 38 (6):1–9.
- Lea, W. 2013. Information: To share or not to share? The Information Governance Review.
- Lindstrand, A., J. Eisfeldt, M. Pettersson, C. M. B. Carvalho, M. Kvarnung, G. Grigelioniene, B.-M. Anderlid, O. Bjerin, P. Gustavsson, A. Hammarsjö, et al. 2019. From cytogenetics to cytogenomics: Whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Medicine 11 (1):1–23. doi: 10.1186/s13073-019-0675-1.
- Lloyd, K., J. McGregor, A. John, N. Craddock, J. T. Walters, D. Linden, I. Jones, R. Bentall, R. A. Lyons, D. V. Ford, et al. 2015. A national population-based e-cohort of people with psychosis (PsyCymru) linking prospectively ascertained phenotypically rich and genetic data to routinely collected records: Overview, recruitment and linkage. Schizophrenia Research 166 (1–3):131–6. doi: 10.1016/j.schres.2015.05.036.
- Lunshof, J. E., and D. Gurwitz. 2013. Guarding children’s genetic privacy. Nature Correspondance 494 (7438):430. doi: 10.1038/494430d.
- Mackley, M. P., B. Fletcher, M. Parker, H. Watkins, and E. Ormondroyd. 2017. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: A systematic review of quantitative and qualitative studies. Genetics in Medicine: Official Journal of the American College of Medical Genetics 19 (3):283–293. doi: 10.1038/gim.2016.109.
- Majumder, M. A., C. J. Guerrini, J. M. Bollinger, R. Cook-Deegan, and A. L. Mcguire. 2017. Sharing data under the 21st century cures act. Genetics in Medicine: Official Journal of the American College of Medical Genetics 19 (12):1289–1294. doi: 10.1038/gim.2017.59.
- Manhas, K. P., S. Page, S. X. Dodd, N. Letourneau, A. Ambrose, X. Cui, and S. C. Tough. 2016. Parental perspectives on consent for participation in large-scale, non-biological data repositories. Life Sciences, Society and Policy 12 (1):1–13. doi: 10.1186/s40504-016-0034-6.
- Manolio, T. A., L. L. Rodriguez, L. Brooks, G. Abecasis, D. Ballinger, M. Daly, P. Donnelly, S. V. Faraone, K. Frazer, S. Gabriel, et al. 2007. New models of collaboration in genome-wide association studies: The genetic association information network. Nature Genetics 39 (9):1045–1051. doi: 10.1038/ng2127.
- Mascalzoni, D., A. Paradiso, and M. Hansson. 2014. Rare disease research: Breaking the privacy barrier. Applied & Translational Genomics 3 (2):23–29. doi: 10.1016/j.atg.2014.04.003.
- Mccabe, L. L., and E. R. B. Mccabe. 2011. Down syndrome: Issues to consider in a national registry, research database and biobank. Molecular Genetics and Metabolism 104 (1–2):10–12. doi: 10.1016/j.ymgme.2011.03.018.
- Mccormack, P., S. Woods, A. Aartsma-Rus, L. Hagger, A. Herczegfalvi, E. Heslop, and J. Irwin. 2013. Guidance in social and ethical issues related to clinical, diagnostic care and novel therapies for hereditary neuromuscular rare diseases: ‘TRANSLATING’ the translational. PLoS Currents 10:5. doi: 10.1371/currents.md.f90b49429fa814bd26c5b22b13d773ec.
- McGowan, M. L., C. A. Prows, M. DeJonckheere, W. B. Brinkman, L. Vaughn, and M. F. Myers. 2018. Adolescent and parental attitudes about return of genomic research results: Focus group findings regarding decisional preferences. Journal of Empirical Research on Human Research Ethics: Jerhre 13 (4):371–382. doi: 10.1177/15562646187766.
- McGuire, A. L., J. M. Oliver, M. J. Slashinski, J. L. Graves, T. Wang, P. A. Kelly, W. Fisher, C. C. Lau, J. Goss, M. Okcu, et al. 2011. To share or not to share: A randomized trial of consent for data sharing in genome research. Genetics in Medicine: Official Journal of the American College of Medical Genetics 13 (11):948–955. doi: 10.1097/GIM.0b013e3182227589.
- McGuire, A. L., M. Basford, L. G. Dressler, S. M. Fullerton, B. A. Koenig, R. Li, C. A. McCarty, E. Ramos, M. E. Smith, C. P. Somkin, et al. 2011. Ethical and practical challenges of sharing data from genome-wide association studies: The EMERGE consortium experience. Genome Research 21 (7):1001–1007. doi: 10.1101/gr.120329.111.
- Mefford, H. C., M. L. Batshaw, and E. P. Hoffman. 2012. Genomics, intellectual disability, and autism. The New England Journal of Medicine 366 (8):733–743. doi: 10.1056/NEJMra1114194.
- Might, M., and M. Wilsey. 2014. The shifting model in clinical diagnostics: How next-generation sequencing and families are altering the way rare diseases are discovered, studied, and treated. Genetics in Medicine: Official Journal of the American College of Medical Genetics 16 (10):736–737. doi: 10.1038/gim.2014.23.
- Nature Biotechnology. 2015. To share is human. Nature Biotechnology 33 (8):796–800. doi: 10.1038/nbt.3309.
- Nooner, K. B., S. J. Colcombe, R. H. Tobe, M. Mennes, M. M. Benedict, A. L. Moreno, L. J. Panek, et al. 2012. The NKI-rockland sample: A model for accelerating the pace of discovery science in psychiatry. Frontiers in Human Neuroscience 6:1–11. doi: 10.3389/fnins.2012.00152.
- Pediatric Imaging. 2011. Neurocognition, and genetics (PING) study.
- Petersen, I., P. Kaatsch, C. Spix, and R. Kollek. 2017. Return and disclosure of research results: Parental attitudes and needs over time in pediatric oncology. The Journal of Pediatrics 191:232–237. doi: 10.1016/j.jpeds.2017.08.008.
- Petrini, C., A. Olivieri, C. Corbetta, R. Cerone, G. D'Agnolo, and A. Bompiani. 2012. Common criteria among states for storage and use of dried blood spot specimens after newborn screening. Annali Dell'Istituto Superiore di Sanita 48 (2):119–121. doi:10.4415/ANN.
- Pinto, N., S. L. Volchenboum, A. D. Skol, L. Rhodes, A. Doan, C. Fein-Levy, J. Lipton, J. M. Cunningham, and K. Onel. 2015. Establishing a translational genomics infrastructure in pediatric cancer: The GREAT KIDS experience. Personalized Medicine 12 (3):221–229. doi: 10.2217/pme.14.90.
- Platt, T., J. Platt, D. Thiel, and S. L. R. Kardia. 2017. Engaging a state: Facebook comments on a large population biobank. Journal of Community Genetics 8 (3):183–197. doi: 10.1007/s12687-017-0302-z.
- Platt, T., J. Platt, D. B. Thiel, N. Fisher, and S. L. R. Kardia. 2014. 'Cool! and creepy': Engaging with college student stakeholders in Michigan's biobank. Journal of Community Genetics 5 (4):349–362. doi: 10.1007/s12687-014-0190-4.
- Rahimzadeh, V., C. Schickhardt, B. M. Knoppers, K. Sénécal, D. F. Vears, C. V. Fernandez, S. Pfister, S. Plon, S. Terry, J. Williams, et al. 2018. Key implications of data sharing in pediatric genomics. JAMA Pediatrics 172 (5):476–481. doi: 10.1001/jamapediatrics.2017.5500.
- Ries, N. M., J. LeGrandeur, and T. Caulfield. 2010. Handling ethical, legal and social issues in birth cohort studies involving genetic research: Responses from studies in six countries. BMC Medical Ethics 11:4. doi: 10.1186/1472-6939-11-4.
- Riggs, E. R., D. R. Azzariti, A. Niehaus, S. R. Goehringer, E. M. Ramos, L. L. Rodriguez, B. Knoppers, H. L. Rehm, C. L. Martin, and On behalf of the Clinical Genome Resource Education Working Group. 2019. Development of a consent resource for genomic data sharing in the clinical setting. Genetics in Medicine: Official Journal of the American College of Medical Genetics 21 (1):81–88. doi: 10.1038/s41436-018-0017-5.
- Robertson, J. A. 2003. The $1000 genome: Ethical and legal issues in whole genome sequencing of individuals. American Journal of Bioethics 3 (3):W–IF1. doi: 10.1162/152651603322874762.
- SACHRP. 2018. Attachment A: Human subjects research implications of ‘big data’. 1–29. https://www.hhs.gov/ohrp/sachrp-committee/recommendations/2015-april-24-attachment-a/index.html.
- Said, M., Ben, L. Robel, B. Golse, and J. P. Jais. 2017. Strengthening data confidentiality and integrity protection in the context of a multi-centric information system dedicated to autism spectrum disorder. Studies in Health Technology and Informatics 245:1133–1137. doi: 10.3233/978-1-61499-830-3-1133.
- Sandelowski, M. 2000. Whatever happened to qualitative description? Research in Nursing & Health 23 (4):334–340. doi: 10.1002/1098-240X(200008)23:4<334::AID-NUR9>3.0.CO;2-G.
- Sanderson, S. C., K. B. Brothers, N. D. Mercaldo, E. W. Clayton, A. H. M. Antommaria, S. A. Aufox, M. H. Brilliant, D. Campos, D. S. Carrell, J. Connolly, et al. 2017. Public attitudes toward consent and data sharing in biobank research: A large multi-site experimental survey in the US. American Journal of Human Genetics 100 (3):414–427. doi: 10.1016/j.ajhg.2017.01.021.
- Schofield, P. N., J. Eppig, E. Huala, M. H. de Angelis, M. Harvey, D. Davidson, T. Weaver, S. Brown, D. Smedley, N. Rosenthal, et al. 2010. Research funding. Sustaining the data and bioresource commons. Science (New York, NY) 330 (6004):592–593. doi: 10.1126/science.1191506.
- Scholtens, S., N. Smidt, M. A. Swertz, S. J. L. Bakker, A. Dotinga, J. M. Vonk, F. van Dijk, S. K. R. van Zon, C. Wijmenga, B. H. R. Wolffenbuttel, et al. 2015. Cohort profile: LifeLines, a three-generation cohort study and biobank. International Journal of Epidemiology 44 (4):1172–1180. doi: 10.1093/ije/dyu229.
- Sofaer, N., and D. Strech. 2011. Reasons why post-trial access to trial drugs should, or need not be ensured to research participants: A systematic review. Public Health Ethics 4 (2):160–184. doi: 10.1093/phe/phr013.
- Strech, D., and N. Sofaer. 2012. How to write a systematic review of reasons. Journal of Medical Ethics 38 (2):121–126. doi: 10.1136/medethics-2011-100096.
- Suresh, S., G. Thangavel, J. Sujatha, and S. Indrani. 2005. Methodological issues in setting up a surveillance system for birth defects in India. The National Medical Journal of India 18 (5):259–262. https://www.scopus.com/inward/record.uri?eid=2-s2.0-30944444199&partnerID=40&md5=9e9c512cb51310f577166ce4ef9e6e88.
- Taylor, D. M., B. J. Aronow, K. Tan, K. Bernt, N. Salomonis, C. S. Greene, A. Frolova, S. E. Henrickson, A. Wells, L. Pei, et al. 2019. The pediatric cell atlas: Defining the growth phase of human development at single-cell resolution. Developmental Cell 49 (1):10–29. doi: 10.1016/j.devcel.2019.03.001.
- Tindana, P., S. Bull, L. Amenga-Etego, J. de Vries, R. Aborigo, K. Koram, D. Kwiatkowski, and M. Parker. 2012. Seeking consent to genetic and genomic research in a rural Ghanaian setting: A qualitative study of the MalariaGEN experience. BMC Medical Ethics 13 (1):15. doi: 10.1186/1472-6939-13-15.
- Wan, Z., Y. Vorobeychik, M. Kantarcioglu, and B. Malin. 2017. Controlling the signal: Practical privacy protection of genomic data sharing through beacon services. BMC Medical Genomics 10 (S2): 39. doi: 10.1186/s12920-017-0282-1.
- Wjst, M. 2010. Caught you: Threats to confidentiality due to the public release of large-scale genetic data sets. BMC Med Ethics 11 (1):21. doi: 10.1186/1472-6939-11-21.
- World Medical Association. 2013. Declaration of Helsinki: Ethical principles for medical research involving human subjects. Amended by the 64 General Assembly, Fotaleza, Brazil.
- Xue, Y. A., Ankala, W. R. Wilcox, and M. R. Hegde. 2015. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: Single-gene, gene panel, or exome/genome sequencing. Genetics in Medicine 17 (6):444–451. doi: 10.1038/gim.2014.122.