Abstract
The presence of small amounts of circulating nucleic acids in plasma and serum (CNAPS) is not a new finding. The verification that such amounts are significantly increased in cancer patients, and that CNAPS might carry a variety of genetic and epigenetic alterations related to cancer development and progression, has aroused great interest in the scientific community in the last decades. Such alterations potentially reflect changes that occur during carcinogenesis, and include DNA mutations, loss of heterozygosity, viral genomic integration, disruption of microRNA, hypermethylation of tumor suppressor genes, and changes in the mitochondrial DNA. These findings have led to many efforts toward the implementation of new clinical biomarkers based on CNAPS analysis. In the present article, we review the main findings related to the utility of CNAPS analysis for early diagnosis, prognosis, and monitoring of cancer, most of which appear promising. However, due to the lack of harmonization of laboratory techniques, the heterogeneity of disease progression, and the small number of recruited patients in most of those studies, there has been a poor translation of basic research into clinical practice. In addition, many aspects remain unknown, such as the release mechanisms of cell-free nucleic acids, their biological function, and the way by which they circulate in the bloodstream. It is therefore expected that in the coming years, an improved understanding of the relationship between CNAPS and the molecular biology of cancer will lead to better diagnosis, management, and treatment.
Video abstract
Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Introduction
According to the World Health Organization, cancer mortality will increase by 45% from 2007 to 2030 due to the demographic increase and population aging.Citation1 For that reason, many efforts have been made to find sensitive and specific biomarkers for early diagnosis, prognosis, and management of patients during treatment and follow-up.
A variety of tumors secrete proteins into the bloodstream that are routinely used as tumor markers in clinical practice, such as the prostate-specific antigen in prostate cancer, the alpha-fetoprotein in hepatocarcinoma, the carcinoembryonic antigen (CEA) in colon cancer, the cancer antigen (CA) 15.3 in breast cancer, and the CA19.9 in pancreatobiliary tumors.Citation2 However, those techniques might lack sensitivity in nonsecretory tumors, and might give positive results due to inflammatory processes or benign illnesses, limiting their specificity. In addition, there are no known serum markers for most tumors, which highlights the need for extensive study of the biology of tumors to propose new clinical tools. Indeed, it is expected that an improvement in the knowledge of the molecular biology of cancer will lead to earlier diagnosis and more effective treatments.
Cancer is produced and progresses as a consequence of complex and gradual processes, in which a variety of genetic and epigenetic alterations are involved (eg, mutations, hypermethylations), and which mainly result in uncontrolled cell growth. Those molecular alterations can be found in the bloodstream,Citation3 which suggests that we could find understanding of the molecular biology of a specific cancer in plasma (). This was the start point for the study of the so-called cell-free nucleic acids in plasma and serum (CNAPS) as cancer biomarkers.
Figure 1 Graphic representation of the analyses of cell-free nucleic acid (cfNA) circulating in plasma that have been tested in cancer patients and that might serve as clinical markers.
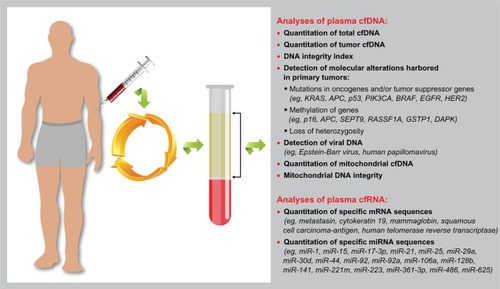
Although in the last decades the CNAPS concept has generated a considerable interest in the research community, the concept was first suggested many years before this. Specifically, Mandel and MétaisCitation4 first reported the existence of cell-free DNA (cfDNA) in plasma in the mid-twentieth century. However, their study had scarce impact until 30 years later, when Leon et alCitation5 demonstrated that cancer patients had higher cfDNA levels than normal people and that patients with persistently high levels of cfDNA after treatment had a worse prognosis than those in whom cfDNA levels decreased.
Some years later, Stroun et alCitation6 suggested that at least some of the cfDNA in serum or plasma was derived from the primary tumor, but this hypothesis was only confirmed later by two nearly parallel studies that described the presence of KRAS mutations in plasma from patients with pancreas neoplasmCitation7 and acute myelogenous leukemia.Citation8 Those discoveries were the milestones that opened new pathways for cancer biology research and the search for new clinical tools.
The term “CNAPS” refers to different types of cell-free nucleic acids (cfNAs), such as genomic DNA (gDNA), mitochondrial DNA (mitDNA), viral DNA and RNA, messenger (m)RNA, and microRNA (miRNA), which have recently been reported to be present in plasma.Citation3,Citation9–Citation13
In spite of the increasing number of studies focused on CNAPS, and of the technological improvements, nowadays, some important aspects of CNAPS biology remain unknown, such as their release mechanisms,Citation14 method of circulation,Citation15–Citation18 and biological role in cancer progression.Citation19 Limiting factors in the study of CNAPS have mainly been the lack of harmonization of laboratory techniques, the heterogeneity of the disease progression, and the small number of recruited patients in most of studies, which is for some authorsCitation9 an example of poor translation of basic research into clinical practice.
Circulating cfDNA
Biology of cfDNA
It has been reported that cfDNA is a double-stranded molecule of low-molecular weight that is fragmented into short (70–200 base pairs) and long sections (up to 21 kilobases).Citation20 The analysis of those fragments has deepened understanding of their origin.Citation21 For some researchers, the main source of cfDNA has been necrosis,Citation22,Citation23 while others have proposed apoptosis in view of the size of the DNA fragments detected in plasma.Citation20 In addition, several studies have suggested the active release of cfDNA by cells,Citation14,Citation24 and specifically by lymphocytes.Citation25 Therefore, two possible sources of cfDNA, non-mutually exclusive, have been considered: passive release through cell death and active release by cell secretion. Nonetheless, recent studies using genome-wide sequencing of plasma DNA have demonstrated that such DNA contains representation of the entire tumor genome and reflects the clonal genomic evolution of tumors.Citation26
Theoretically, CNAPS would be rapidly degraded in the bloodstream by nucleases; it has even been proven that mutated cfDNA degrades faster than non-mutated cfDNA.Citation27,Citation28 However, the enzymatic action might be limited because at least part of cfDNA appears to be protected by being complexed or particulate with special protective characteristics against enzymatic degradation.Citation29–Citation31 A decreased activity of DNase has also been observed in plasma from cancer patients, which might be another reason for the high levels of cfDNA found in plasma.Citation31
Quantitation of cfDNA in plasma and serum
Many studies have confirmed the early findings of Leon et al.Citation3,Citation9 In fact, high concentrations of cfDNA have been reported in plasma and serum from patients with various cancers, such as cancer of the colon,Citation33 lung,Citation34–Citation36 breast,Citation37,Citation38 stomach,Citation39 and esophagus.Citation40 The clinical value of such quantitation for differential diagnosis was suggested in a large study by Shapiro et al,Citation41 who showed that patients with benign gastrointestinal diseases had a lower mean concentration of plasma cfDNA (118 ng/mL) than cancer patients (412 ng/mL).
Several studies have proposed the use of the quantitation of cfDNA in plasma as a method for screening colon,Citation33 breast,Citation38 and lung cancersCitation36,Citation42 among others. In colon cancer patients, such a method has showed an even better sensitivity than CEA quantitation.Citation33 In breast cancer patients, plasma levels of cfDNA have also been related to clinicopathological variables such as size, tumor stage, lymphadenopathies, human epidermal growth factor receptor 2 (HER2)/neu level and state.Citation38 In lung cancer, it has even been suggested that the concentration of circulating DNA might be a risk factor for the presence of the illness and a prognostic index during follow-up.Citation36
However, in extensive revisions, the use of quantitation of cfDNA in plasma as a unique marker has been questionedCitation9 and has even been proposed as inadvisable in lung cancerCitation43 and associated with CA125 in ovarian cancer.Citation44 In addition, it is remarkable that overlapping concentrations of cfDNA are found in healthy individuals under physiological stress (eg, physical exercise) or in patients affected by other pathological processes, such as, inflammation, trauma, or sepsis.Citation3,Citation45–Citation47
The validation of the clinical utility of methods for detection and quantitation of plasma cfDNA has probably failed because of technical limitations, particularly those related to sensitivity and specificity. There have been some attempts to establish reference values for different types of cancer;Citation48 however, the lack of harmonization in the laboratory techniques (quantitative polymerase chain reaction [PCR], spectrophotometry, fluorimetry, etc) and the low number and heterogeneity of patients enrolled in each study, have prevented the achievement of suitable statistical power and the establishment of reference patterns.Citation3,Citation9,Citation10
Although, theoretically, the levels of cfDNA in plasma might be affected by several clinicopathological features such as tumor size, tumor stage, or metastasis,Citation9,Citation38 no direct relationship between these features has been proven, as Lecomte et al discussed in their review focused on colorectal cancer.Citation49 García-Olmo et al have conducted studies in animal models to deepen understanding of cfDNA kinetics and have repeatedly shown that non-mutated DNA levels are not significantly related to tumor size or metastasis.Citation50,Citation51 In fact, they found that large amounts of non-tumor DNA are released during tumor progression and, in particular, at the early stages, suggesting that there is active interaction between tumor and non-tumor cells.Citation50
It is probable that the quantitation of cfDNA in plasma will be of most clinical value during disease monitoring.Citation42 It has been reported that plasma levels decreased in cancer patients after surgical treatmentsCitation33,Citation35,Citation37,Citation39,Citation40 and/or chemoradiotherapy,Citation52 sometimes reaching levels similar to those measured before treatment.Citation35,Citation39,Citation40 In addition, patients who maintained high levels of cfDNA in plasma either did not respond to the treatment or had a high risk of relapse.Citation5,Citation33–Citation37,Citation53
Circulating nucleosomes, as degradation products of necrotic tumor cells in the bloodstream, have also been examined and quantified by enzyme-linked immunosorbent assay techniques.Citation16 The DNA of necrotic cells, after being phagocytosed by macrophages,Citation23 might be released to the bloodstream inside those structures, which protect it from enzymatic degradation. Nucleosomes have been found in healthy subjects and in patients with benign diseases,Citation15 thus, their value as a screening method appears to be limited; however, there is some evidence to support their utility for cancer monitoring. Specifically, during chemotherapy treatments, an initial increase of the levels of nucleosomes has been observed in plasma, which might be related to apoptosis provoked by treatment, and a later decrease in patients who had a good response to the treatment.Citation15,Citation54,Citation55
Molecular alterations of cfDNA
Most of the molecular alterations found in cfDNA circulating in plasma reflect the genetic and epigenetic changes found in primary tumors and, thus, the analysis of such tumor cfDNA might be valuable for tumor diagnosis and monitoring. Highly sensitive methods are required to detect those alterations among larger quantities of non-altered cfDNA molecules and, for this reason among others, extremely varied results have been reported.Citation3,Citation20,Citation56
Following, we review the alterations in cfDNA most frequently found in plasma from cancer patients and tumor-bearing animals.
Integrity of the DNA strand
Using PCR, Wang et alCitation57 found that long DNA fragments, related to necrosis phenomena, could be distinguished from shorter fragments, produced by physiological apoptosis phenomena. These findings gave rise to the so-called integrity index, based on the ratio between long and short cfDNA fragments. The integrity index was established based on gynecologic and breast cancers,Citation57 with the results obtained from 61 patients compared with those from 65 patients without neoplastic disease. It was found that the area under the curve for the DNA integrity index was 0.911 in cancer patients in relation to patients without cancer, with 100% sensitivity and 62% specificity. The authors suggested that the integrity index provides a simple and inexpensive way to detect cancer.Citation57
A number of transposable elements of the genome, such as ALU and LINE1 sequences, can be easily detected and have been associated with tumor necrosis. Specifically, ALU sequences are short elements (typically 300 nucleotides in length) that account for more than 10% of the human genome. Umetani et al developed a method to measure the integrity of cfDNA in serum using quantitative PCR for ALU repeats, and suggested that the integrity index is increased at the early stages of the disease and a promising molecular biomarker for detecting colorectal and breast cancer.Citation58,Citation59 Moreover, it might serve as a sensitive method to detect nodal metastases in the early stages, having an even higher predictive value than the clinicopathological variables commonly used, such as tumor size and stage.Citation59
In a recent study in patients with hepatitis B virus-related hepatocellular carcinoma, the integrity index was associated with tumor size, TNM Classification of Malignant Tumors (TMN) stage, and nodal metastasis, and showed a more discriminatory power than total cfDNA concentration.Citation60
Further, in other recent studies, the clinical value of the integrity index has been demonstrated, not only for the diagnosis but also the monitoring of the disease. For example, it has been found that the integrity index decreases in parallel to the response to surgical treatments for head and neck tumorsCitation61 and melanoma,Citation62 to chemotherapy in leukemia,Citation63 to radiotherapy in nasopharyngeal tumors,Citation64 and to chemoradiotherapy in colorectal cancer.Citation52 Moreover, it has been reported that patients with high concentrations of integrity DNA had shorter disease-free intervals.Citation64
Alterations in the integrity of DNA have also been detected in other organic liquids, such as urine, and can be used in the early diagnosis of bladder cancer without cytology. Citation65 Some authors have emphasized that the integrity index meets many of the requirements of a universal biomarker;Citation10 however, other studies have not supported this idea.Citation66
Gene mutations
As previously mentioned, many studies have shown that gene mutations found in primary tumors can be also detected in cfDNA circulating in plasma. The literature reports a wide percentage range of patients in whose plasma such mutations have been detected. The differences between studies might be due to technical reasons, since sensitive techniques are needed to detect very small amounts of mutated DNA among larger quantities of un-mutated DNA, which has a dilution effect.
The gene that has most frequently been examined is the KRAS oncogene, which is mutated in variable rates in high-incidence tumors, such as colon, pancreas, lung and thyroid tumors,Citation9 and has intrinsic characteristics that make it useful as a marker.Citation49 In fact, the prevalence of KRAS mutations is near 50%. Such mutations occur in the early stages of carcinogenesis and are mostly produced at specific sites (particularly in codon 12), which facilitate its detection using PCR-based techniques.
With respect to colorectal cancer, KRAS mutations have been found in 40%–50% of patients, and have also been detected in the plasma or serum of 25%–30% of patients.Citation49 Kopreski et alCitation67 tried to correlate the detection of KRAS mutations in plasma with clinicopathological findings detected by colonoscopy. They enrolled into their study one of the largest series of colorectal cancer patients that has ever been enrolled into similar studies, and detected KRAS mutations in plasma from 83% of patients whose tumors had such mutations.Citation67 Moreover, they also found that some patients with apparently normal colonoscopy had KRAS mutations in plasma, which might be related to the presence of precancer lesions, thus suggesting the technique’s suitability for early diagnosis and screening.Citation67
In a review by Sorenson,Citation68 the author defended the detection of KRAS mutations in plasma as a specific marker for gastrointestinal tumors, with 2.5% false positives, although the review of studies showed that the concordance with KRAS plasmatic mutations with respect to primary tumor was only 50%. It has also been reported that other mutations, different from those in primary tumors, can be detected in plasma from cancer patients, which might be due to clone heterogeneity in tumors.Citation69
In patients with pancreas cancer, it has been shown that detecting KRAS mutations in plasma is useful for early diagnosis as a complementary marker together with other serum markers (eg, CA19.9).Citation70 Other researchers have also used it for disease monitoring.Citation71
In a prospective study conducted by Gormally et alCitation72 of a healthy population, KRAS mutations were detected in plasma from 1.2% subjects and p53 mutations in 3.6%. The authors suggested that KRAS mutations are detectable in plasma before the diagnosis of bladder cancer. In contrast, KRAS mutations have been detected by other researchers in up to 30% of healthy individuals;Citation9 this has been interpreted as a limitation of the diagnosis and screening value.
With respect to mutations in other genes, Chen et alCitation73 used an ultrasensitive technique to detect p53 mutations in plasma from patients with Stage II and III breast cancer, and their results showed a correlation between such detection and the clinical course of the disease after therapy.
The value of detection of tumor mutations in plasma for monitoring the response to treatment has been analyzed in several studies. Diehl et alCitation74 showed that the detection and quantification of mutated cfDNA in plasma from colon cancer patients undergoing surgery and chemotherapy were more useful for monitoring than the quantitation of CEA in serum. In addition, a recent study has shown that the use of panels with the most frequent mutations in colorectal cancer (APC, KRAS, TP53, PIK3CA, and BRAF) was more useful than using CEA and CA19.9 levels.Citation75 Further, it has been suggested that, in the postoperative period, the levels of mutated KRAS in plasma are a more powerful predictor of recurrence than Dukes stage.Citation76
In breast cancer patients, the presence of amplified HER2 has been demonstrated in circulating cfDNA during follow-up.Citation77 This led the researchers to propose such amplification as a marker for prognosis and response to treatment with monoclonal antibodies, such as trastuzumab.Citation77 A recent study has provided proof of the concept that tumor cfDNA circulating in plasma represents a highly sensitive biomarker of tumor burden in metastatic breast cancer.Citation78 Specifically, the researchers developed new methods to identify somatic genomic alterations (point mutations, structural variants) and designed personalized assays to quantify tumor cfDNA circulating in plasma from 30 patients. They found that levels of tumor cfDNA showed a greater dynamic range and greater correlation with changes in tumor burden than did CA15-3 or circulating tumor cells.Citation78
Many chemotherapeutic agents act on pathways in which KRAS, BRAF, EGFR, or p53 are involved.Citation79,Citation80 For this reason, in many cases, it is important to know the mutation status for predicting the response to treatment and monitoring the disease. The analysis of plasma offers a noninvasive and quick way to find out this information and, in this sense, it has been reported that detection of EGFR mutations in plasma might be useful to predict disease progression, disease-free intervals, and drug resistances in patients with lung cancer.Citation79–Citation81 Moreover, it has recently been reported that sequencing of cancer exomes in serial plasma samples might be useful to track genomic evolution of metastatic cancers in response to therapy.Citation26 The researchers described a noninvasive approach for characterizing cancer exomes in plasma that might enable detailed and comprehensive evaluation of clonal genomic evolution associated with treatment response and resistance.Citation26
Summarizing, patients whose tumors have specific mutations might be monitored by analyzing the tumor cfDNA in their plasma samples.Citation10,Citation26,Citation78
Gene hypermethylations
Some tumors are related to specific epigenetic alterations (eg, methylation), leading to changes in the expression of promoters of suppressor genes, which results in their silencing. Such alterations occur early in tumorigenesis and in DNA fragments that are rich in cytosine and guanine (CpG islands). The first observations were reported in 1999 by Esteller et alCitation82 in non-small cell lung carcinoma; now, the detection of these alterations represents one of the most promising advances in cancer diagnosis.
Epigenetic alterations are not tumor specific; moreover, there are some genes that are frequently hypermethylated and silenced in different types of tumors. In fact, many studies have analyzed panels of genes to increase sensitivity.Citation83–Citation85 Thus, it is essential to select accurately the genes to analyze for each type of cancer to improve the sensitivity of the analyses. Some of the most common aberrant methylations affect the p16 tumor suppressor gene and have been found in patients with liver, lung, and breast tumors.Citation82,Citation86,Citation87 Other suppressor genes frequently hypermethylated are SEPT9, RASSF1A, GSTP1, and DAPK, among others.
The clinical value of the detection of hypermethylated genes in plasma has been shown in different types of cancer such as breast,Citation84,Citation85 colon,Citation53 liver,Citation88 esophagus,Citation89 and urological tumors,Citation90 as well as in hematologic diseases.Citation91 It has been suggested that the methylated status of cfDNA circulating in plasma might be a tool for prognosis stratification and the prediction of the response to some chemotherapeutics agents,Citation8,Citation9,Citation92 disease-free interval, and risk of relapse.Citation53,Citation88,Citation89
Hypermethylations have also been detected in cfDNA from other body fluids such as urine.Citation90 Specifically, the presence of tumor cfDNA has been reported in 70% of urine samples, suggesting its utility for the diagnosis and monitoring of patients with urologic tumors such as bladder, prostate and kidney cancers.Citation90
Microsatellite alterations
“Microsatellites” are repeating sequences of one to six nucleotides that are scattered along the genome; their function is unknown. Such sequences serve to identify “loss of heterozygosity” (LOH), which is a frequent alteration of tumor DNA characterized by the loss of an allele when compared with matched normal DNA from the same individual. LOH indicates the absence of a functional tumor suppressor gene by deletion. In contrast, some tumors have abnormally long or short microsatellites as a result of a defective DNA repair process; this is termed “microsatellite instability” (MSI).
It has been reported that LOH and MSI can be detected in cfDNA circulating in plasma; however, discrepancies between tumor DNA and plasma cfDNA have been reported.Citation2 Although the detection of such alterations in plasma is more probable in advanced stages, some researchers have suggested a potential value for such detection at the diagnosis stage of breastCitation93 and ovarian cancers,Citation44 which may have an even higher sensitivity than the quantitation of cfDNA.Citation44
To increase the sensitivity of this kind of marker, they have been tested in combination with several other plasma markers, such as methylations and prostate-specific antigen in prostate cancer.Citation94 In addition, panels of microsatellites are often used to improve sensitivity.Citation95,Citation96 For example, in a recent prospective study in breast cancer patients, LOH was determined by PCR-based microsatellite analysis using a panel of eight polymorphic markers.Citation96 The researchers found that LOH at those markers was significantly correlated with tumor stage, tumor size, lymph node metastasis, positive progesterone, and HER2 status.Citation96 Moreover, LOH at a marker mapping to cyclin D2 correlated with shorter overall survival. Thus, the researchers concluded that the improved detection of LOH on cfDNA provides important information on DNA losses of tumor suppressor genes (TIG1, PTEN, cyclin D2, RB1, and BRCA1) in breast cancer. In particular, loss of the cyclin D2 gene may become an important prognostic marker easily detectable in the peripheral blood.Citation96
Prior to that interesting study, it was reported that the assessment of microsatellite status in plasma might be a useful predictive tool for prognosis in breast carcinoma,Citation97 to monitor the response to surgical treatment; in oral squamous cell carcinoma;Citation95 and to biochemotherapy in metastatic melanoma.Citation98
Viral DNA
Viruses are the main etiologic factors of a number of tumors and can be detected in plasma by PCR-based techniques. In fact, the presence of cfDNA of Epstein–Barr virus (EBV) has been demonstrated in nasopharyngeal cancers, Hodgkin’s disease, and Burkitt lymphoma; human papillomavirus in cervical tumors; and hepatitis B virus in hepatocellular tumors, certain lymphomas, and gastric cancers.Citation3,Citation10,Citation12,Citation99–Citation102
The presence of EBV sequences in plasma has been the focus of many studies, perhaps due to the high incidence of nasopharyngeal carcinoma in Asian countries. It has been shown that the detection of EBV in plasma is a powerful diagnostic tool and its quantitation might have prognostic value.Citation103 In addition, detection of EBV in plasma has also been associated with response to radiotherapy, disease recurrence, and survival.Citation12
mitDNA
Although the study of the genetic and epigenetic alterations of gDNA is the cornerstone of cancer research, the discovery of specific alterations in mitDNA in cancer patientsCitation104 has opened new routes in the search for clinical tools. The detection of aberrant changes in mitDNA is becoming an important tool for the early diagnosis of cancer, which is in part due to the fact that the analysis of mitDNA has some advantages over that of gDNA.Citation11 These advantages can be summarized as follows.
The mitochondrial genome is shorter and more simply organized than nuclear DNA. These unique properties make the screening of a mitochondrial genome much easier and more cost-effective.
mitDNA’s high number of copies, in comparison to nuclear DNA, make it a much more sensitive method.
mitDNA fragments have been detected in different body fluids from cancer patients at early stages, such as in the bloodCitation104,Citation105 saliva,Citation106 urine,Citation107 and sputum.
The alteration of mitDNA as a response to adaptation changes was first described in the early twentieth century, when it was termed the “Warburg effect.”Citation108 Alterations in mitDNA have been found in the plasma of healthy individualsCitation109 and have also be related to tumor development and progression.Citation110 The alterations, which are point mutations, deletions, insertions, and quantitative changes,Citation11 have been detected in a wide range of tumors, such as breast, colon, liver, head and neck and lung.Citation111 It is important to highlight that the identification of mutations in a concrete region typical to many tumors is referred to as the “D-loop,” which may have diagnostic value.Citation104
The presence of mitDNA mutations has been reported in plasma and serum samples from patients with hepatocellular, pancreatic, prostatic, colorectal, and esophageal cancers, among others.Citation112–Citation116 Moreover, several studies have suggested that quantitation of mitochondrial cfDNA in plasma might serve as a clinical tool,Citation117,Citation118 and may even have a higher diagnostic value than gDNA in some cases.Citation118 Specifically, Kohler et alCitation117 compared the levels of gDNA and mitDNA in plasma from patients with benign and malignant breast tumors with those from healthy controls. They concluded that both nuclear and mitochondrial cfDNA have potential as biomarkers in breast tumor management; however, the nuclear cfDNA showed greater sensitivity and specificity. Zachariah et alCitation118 conducted a study in 104 women with ovarian cancer, benign tumors, and endometriosis, and compared the levels of gDNA and mitDNA in plasma in these patients. Patients with epithelial ovarian cancer had significantly higher amounts of nuclear and mitochondrial cfDNA than the other women, but the levels of cfDNA in plasma were related to neither pathological parameters nor CA125 levels. In addition, the researchers found that quantitation of mitDNA was a unique way to differentiate between patients with ovarian cancer and endometriosis.Citation118
Other studies have used “mitochondrial DNA integrity,” which has been defined as the ratio between long and short fragments of mitDNA.Citation119 It has been reported that such an index might differentiate between patients with urologic malignancies (renal, prostate, and bladder tumors) and healthy subjects, with a sensitivity of 84% and a specificity of 97%.Citation119
Finally, it has been reported that some mitDNA polymorphisms are associated with cancer development, thus a genetic analysis of such polymorphisms could help to identify target populations to establish screening programs.Citation11
Circulating cell-free RNA (cfRNA)
Biology of cfRNA
The origin of the cfRNA circulating in plasma, its role, and its release mechanisms are yet unknown. The existence of cfRNA in blood was reported many years ago when, in 1987, Wieczorek et alCitation120 found RNA in proteolipid complexes in the serum of cancer patients.Citation121 However, the potential clinical value of the detection of cfRNA in plasma did not attract the attention of researchers until 1999, when two parallel studies reported the detection of tyrosinase mRNA in patients with metastatic melanomaCitation122 and mRNA associated with EBV in patients with nasopharyngeal carcinoma.Citation123 Many studies have subsequently reported the presence of specific mRNA in plasma from patients with a variety of cancers including colon,Citation124–Citation126 breast,Citation127,Citation128 prostate,Citation129,Citation130 melanoma,Citation122 lung,Citation131 and thyroid.Citation132
It was theorized that the fragility of cfRNA in serum or plasma – due to the fast enzymatic degradation it undergoes, which is increased in cancer patientsCitation133 – might make its detection difficult. However, it was found that the molecules are more stable than it was presumed,Citation134 possibly due to protection by vesicle-like structures.
The origin of cfRNA remains less clear than that of cfDNA.Citation21,Citation24 It has been proposed that apoptosis might be involved in the release of cfRNA, and that its association with apoptotic bodies might explain the resistance to nucleases in blood.Citation31 However, other hypotheses for the circulation of cfRNA have also been put forward, such as it occurring within lipoprotein complexesCitation24 or being in other actively-released particles, such as exosomes.Citation121
Ng et alCitation135 examined the particle-associated nature of circulating cfRNA by filtering plasma samples from healthy subjects and cancer patients through material with different pore sizes. They found greater amounts of particle-associated mRNA in cancer patients than in healthy subjects, suggesting that most of the cfRNA not associated with particles had degraded.Citation135
Detection and quantitation of cfRNA in plasma and serum
The use of cfRNA as a biomarker has several advantages including the ease with which plasma or serum samples for testing can be obtained, which makes it feasible for the monitoring of metastatic disease and even for wide screenings. However, RNA molecules are fragile, so high sensitivity techniques with simultaneous internal controls are necessary.Citation136
One of the most analyzed mRNAs in plasma is that of the human transcriptase reverse telomerase (hTERT), which corresponds to a ribonucleoprotein involved in the repair and lengthening of telomeres in eukaryotic cells. This mRNA is overexpressed in a variety of tumors.Citation124,Citation126,Citation128,Citation129,Citation137–Citation140 However, cfRNA is not specific to cancer patients and can also be found in healthy volunteers or in those suffering trauma.Citation46,Citation141
In breast cancer patients, using PCR-based techniques, hTERT mRNA has been detected in serum, even in patients with localized disease.Citation128 Silva et alCitation127 observed that analysis of specific mRNA epithelium (cytokeratin 19) in plasma, in combination with mammaglobin, facilitates the detection of a greater number of positive cases than does analysis of tumor cfDNA (73% versus [vs] 29%). Moreover, detection of epithelial mRNA has been found to be related to tumor size and proliferation rate.Citation127 In addition, El-Abd et al have suggested the utility of the detection of metastasin mRNA in serum as a survival marker, with high sensitivity (85%) and specificity (100%).Citation142
In colon cancer patients, an adequate correlation between hTERT levels in plasma and tumor stage has been observed, which has led such a quantitation to be proposed as a tool for screening, monitoring,Citation137 and response to treatment.Citation140
With respect to lung cancer, Miura et alCitation138 analyzed plasma samples from 112 patients and 80 healthy subjects to detect and quantify hTERT and epidermal growth factor receptor (EGFR) mRNAs. They found that the sensitivity and specificity in lung cancer diagnosis were, respectively, 89% and 73% for hTERT mRNA, and 71% and 80% for EGFR mRNA. Moreover, they found that the number of copies of hTERT mRNA significantly decreased after surgical treatment.Citation138 These data led the researchers to suggest that hTERT mRNA, especially when combined with EGFR mRNA, may be an excellent biomarker for pulmonary malignancies to diagnose and assess clinical stage.Citation138 In addition, it has been suggested that the detection of heterogeneous nuclear ribonucleoprotein-B1 mRNA and HER2/neu-specific mRNA might have diagnostic value in lung cancer.Citation143
In esophagus cancer patients, the detection of squamous cell carcinoma-antigen mRNA (SCC-Ag mRNA) by realtime PCR was shown to be the best predictive factor for recurrence in patients.Citation144
In hepatocellular cancer, the detection of hTERT mRNA in serum has been suggested to be of diagnostic value, with a sensitivity of 88% and a specificity of 70%.Citation139 Moreover, the researchers reported a good correlation between the levels of hTERT mRNA in plasma and clinicopathological parameters, such as degree of differentiation.Citation139 However, other studies have shown no correlation with clinicopathological variables such as tumor size.Citation145
cfRNA can be detected in other body fluids such as saliva and urine, and satisfactory results have been reported for its detection in these as a marker for the diagnosis of mouth and urological cancers, respectively.Citation136
miRNA
The discovery of miRNA in 1993 was a milestone in cell biology research. From that moment, the number of identified miRNAs steadily increased and, to date, more than 900 human miRNAs have been discovered.
“miRNAs” are small noncoding molecules of 18–22 nucleotides that regulate gene expression, acting at a post-transcriptional level. It is known that a unique miRNA is able to regulate multiple different mRNAs, so that it might potentially alter the function of multiple channels.Citation13
miRNAs have been demonstrated to play important roles in control of cell proliferation, cell differentiation, and apoptosis, thus their dysregulation contributes to tumor development. Moreover, it has been demonstrated that miRNAs can function as potential oncogenes or oncosuppressor genes, depending on the cellular context and on the target genes they regulate.Citation146 Aberrant expression of miRNAs has been associated with multiple types of cancer including colon,Citation147–Citation150 lung,Citation151–Citation154 breast,Citation155 prostate,Citation156 gastric,Citation157 and esophagus,Citation158 as well as lymphoma.Citation159
miRNAs have been detected in the plasma and serum of animals and humans, and in both healthy subjects and cancer patients. The first evidence that miRNAs might serve as serological biomarkers of solid tumors was provided by Mitchell et alCitation156 and Chen et al.Citation147 miRNAs circulating in plasma have some advantages as clinical markers compared with other forms of cfRNA, since they have a remarkable resistance to endogenous and exogenous ribonuclease activity, extreme pH conditions, and freeze–thaw cycles, whereas synthetic miRNAs are promptly degraded.Citation160
Circulating miRNAs have been found packaged into various membrane-bound vesicles such as exosomes,Citation18 microvesicles and apoptotic bodies (reviewed by Zandberga et al),Citation160 and also in lipoprotein complexes.Citation141 This is probably the main reason for the high resistance of plasma miRNAs, which makes their detection by PCR-based techniques easy. In addition, miRNAs are tissue specific, which allows the development of panels to help in the diagnosis of tumors of unknown origin.Citation156,Citation161,Citation162 Moreover, they are not only detectable in the bloodstream but also in other body fluids.Citation18,Citation163 Thus, the use of miRNAs as serological biomarkers is a very attractive option and research in this field continues to increase.
miR-21, probably the most analyzed miRNA, is often overexpressed in many types of tumors.Citation164 Nonetheless, a number of miRNAs have been demonstrated to have potential value as serological biomarkers in cancer, including miR-1, miR-10b, miR-17-92, miR-24, miR-92a, miR-122, miR-141, miR-155, miR-195, miR-221, and miR-375.Citation165
The diagnostic value of miRNA panels appears to be superior to that of individual miRNAs.Citation160 Specific expression patterns of serum miRNAs have been identified for lung cancer, colorectal cancer, and diabetes, providing evidence that serum miRNAs contain fingerprints for a variety of diseases.Citation147
It has been demonstrated that colorectal cancer patients have an miRNA serum profile significantly different to that of healthy subjects. Specifically, 69 miRNAs have been detected in sera from colorectal cancer patients but not in those from healthy subjects.Citation147 Moreover, colorectal cancer patients have been found to share a large number of serum miRNAs with lung cancer patients.Citation147 In addition, Ng et alCitation148 analyzed plasma samples from 90 colorectal cancer patients and 50 healthy subjects and found that miR-17-3p and miR-92 were overexpressed in patients, which suggested the potential diagnostic value of such plasma levels, with a sensitivity of 89% and specificity of 70%. Subsequent studies have supported Ng et al’s findings, including that by Huang et al,Citation149 who showed that levels of miR-29a and miR-92a in plasma discriminate colorectal cancer, with 83% sensitivity and 85% specificity. In a recent revision, the prediction value of three miRNAs (miR-221, miR-141, and miR-29a) has been highlighted as 3 independent factors of wrong predictions in different clinical stages.Citation166
The increased expression of miR-21, miR-106, and miR-15 has been observed in breast cancer patients compared with that observed in healthy subjects, in both tissues and serum, and those levels have been found associated with tumor stage and the presence of lymph node metastases.Citation155 With respect to lung cancer, Wang et alCitation152 and Liu et alCitation153 observed high levels of miR-21 in the plasma of cancer patients compared with that in healthy individuals. These levels were also associated with TNM stage and the presence of lymph node metastasis. Further, Roth et alCitation154 suggested that miR-361-3p and miR-625 might have a protective effect on the development of non-small cell lung cancer, while its quantification in serum might have the diagnostic potential to detect that cancer, particularly in smokers.
The most comprehensive search to date for circulating miRNAs with prognostic significance was performed by Hu et al,Citation151 who analyzed a large cohort of 303 patients and found that four plasma miRNAs (miR-486, miR-30d, miR-1, and miR-449) were independent predictors of average survival. In prostate cancer, Mitchell et alCitation156 were able to distinguish patients with cancer from healthy individuals by serum levels of miR-141. Finally, Lawrie et al demonstrated that serum levels of miR-21 were associated with the disease-free interval in patients with diffuse B-cell lymphoma.Citation159
In future, the search for circulating miRNAs of clinical value is likely to be carried out at the same time as the study of many unknown aspects, such as the role of circulating miRNAs, the cell types that secrete them, and the regulation of this secretion.
Conclusion
The data reviewed in the present article suggest that the clinical value of cfNAs circulating in plasma is already more than a theoretical idea, since the characterization and the quantitation of such nucleic acids (NAs) have been shown to be complementary tools in the diagnosis, prognosis, and management of cancer patients (). However, some important questions remain unanswered, largely because most studies have been underpowered as well as because there has been no adequate standardization of laboratory techniques, which complicates the comparison of results from different groups.Citation3,Citation9,Citation10 Thus, it appears necessary to drive initiatives directed toward validating and verifying laboratory methods and procedures for required molecular tests before their use in clinical testing. For this, it might be useful to network, as has been demonstrated by successful previous initiatives.Citation167 Methods based on sequencing (genome-wide sequencing, exome sequencing, etc) appear to be the definitive tool for making the analysis of cfDNA for cancer monitoring useful, since it allows for personalized assays.Citation26,Citation78
Figure 2 Schematic representation of the outline of the present review.
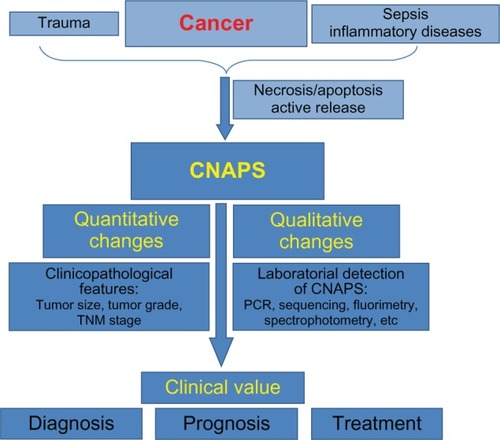
However, plasma cfNAs have yet many enigmatic aspects. Elucidation of these might lead us to a revision in the way the pathobiology of cancer is conceived. It is likely that many molecular pathways are involved in the origin of such NAs, since various results point in non-exclusive different directions. It has been postulated that the cfNAs circulating in plasma are not merely biological waste, but that they may be directly involved in the development of metastases, possibly through transfection-like uptake by susceptible cells.Citation168 This hypothesis, the “genometastasis theory,” is supported by strong evidence – namely, the observation that plasma from cancer patients can transfect and oncogenically transform cultured cells.Citation169,Citation170
Regardless, no conclusive explanation of the circulation mechanism of cfNAs yet exists. Several groups have suggested a relationship between the presence of cfNAs in plasma and the presence of exosomes.Citation121,Citation171 Thus, it appears feasible that at least part of plasma cfNAs circulates within exosomes. This finding correlates with the idea that plasma cfNAs might have a role in tumor progression, since it has been proven that exosomes are able to transfer their RNA content to cells, and that this RNA can be functional in its new location.Citation18 It has even been demonstrated that miRNAs are transferred during immune synapsis and are able to modulate gene expression in recipient cells.Citation172 Moreover, cross-talk between tumor-derived exosomes and host cells, such as bone marrow progenitor cells, appears undeniable.Citation173 Thus, transfer of genetic material from exosomes to cells might be involved in the recruitment and metastatic conversion of host cells. Conversely, it might be possible to exploit this phenomenon for therapeutic purposes and, perhaps, it will be proper to focus the development of clinical tools on the detection and analysis of NAs containing microparticles. Certainly, at this moment, a robust body of research on exosomes and microvesicles exists and increased understanding of such particles in future will shed light on cancer pathobiology and offer promising perspectives on clinical tools.
Thus, although the origin and the circulation mechanism of plasma cfNAs remain unclear, there is strong evidence to suggest that such NAs will become useful biomarkers for the diagnosis and monitoring of cancer disease and that they might be essential to tumor development and progression.
Acknowledgments
The authors thank Mr Eduard Cervera for his invaluable help in designing .
Disclosure
The authors declare no conflicts of interest in this work.
References
- World Health Organization [homepage on the Internet] Available from: http://www.who.int/features/qa/15/en/Accessed June 6, 2013
- JacobsELHaskellCMClinical use of tumor markers in oncologyCurr Probl Cancer19911562993601760927
- FleischhackerMSchmidtBCirculating nucleic acids (CNAs) and cancer – a surveyBiochim Biophys Acta20071775118123217137717
- MandelPMétaisPLes acides nucléiques du plasma sanguin chez l’homme [The nucleic acids of blood plasma in humans]Compte Rendu de l’Academie des Sciences1948142241243 French
- LeonSAShapiroBSklaroffDMYarosMJFree DNA in the serum of cancer patients and the effect of therapyCancer Res1977373646650837366
- StrounMAnkerPMauricePLyauteyJLederreyCBeljanskiMNeoplastic characteristics of the DNA found in the plasma of cancer patientsOncology19894653183222779946
- SorensonGDPribishDMValoneFHMemoliVABzikDJYaoSLSoluble normal and mutated DNA sequences from single-copy genes in human bloodCancer Epidemiol Biomarkers Prev19943167718118388
- VasioukhinVAnkerPMauricePLyauteyJLederreyCStrounMPoint mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemiaBr J Haematol19948647747797918071
- JungKFleischhackerMRabienACell-free DNA in the blood as a solid tumor biomarker – a critical appraisal of the literatureClin Chim Acta201041121–221611162420688053
- SchwarzenbachHHoonDSPantelKCell-free nucleic acids as biomarkers in cancer patientsNat Rev Cancer201111642643721562580
- YuMSomatic mitochondrial DNA mutations in human cancersAdv Clin Chem2012579913822870588
- TsangJCLoYMCirculating nucleic acids in plasma/serumPathology200739219720717454749
- GiovannettiEErozenciASmitJDanesiRPetersGJMolecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practiceCrit Rev Oncol Hematol201281210312221546262
- GahanPBAnkerPStrounMMetabolic DNA as the origin of spontaneously released DNA?Ann N Y Acad Sci2008113771718837918
- HoldenriederSStieberPBodenmüllerHCirculating nucleosomes in serumAnn N Y Acad Sci20019459310211708501
- HoldenriederSStieberPBodenmüllerHNucleosomes in serum as a marker for cell deathClin Chem Lab Med200139759660511522104
- IguchiHKosakaNOchiyaTSecretory microRNAs as a versatile communication toolCommun Integr Biol20103547848121057646
- ValadiHEkströmKBossiosASjöstrandMLeeJJLötvallJOExosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cellsNat Cell Biol20079665465917486113
- García-OlmoDCGarcía-OlmoDBiological role of cell-free nucleic acids in cancer: the theory of genometastasisCrit Rev Oncog2013181–215316123237557
- JahrSHentzeHEnglischSDNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cellsCancer Res20016141659166511245480
- StrounMLyauteyJLederreyCOlson-SandAAnkerPAbout the possible origin and mechanism of circulating DNA apoptosis and active DNA releaseClin Chim Acta20013131–213914211694251
- LiCNHsuHLWuTLTsaoKCSunCFWuJTCell-free DNA is released from tumor cells upon cell death: a study of tissue cultures of tumor cell linesJ Clin Lab Anal200317410310712784257
- ChoiJJReichCFPisetskyDSThe role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cellsImmunology20051151556215819697
- StrounMMauricePVasioukhinVThe origin and mechanism of circulating DNAAnn N Y Acad Sci200090616116810818614
- ThijssenMASwinkelsDWRuersTJde KokJBDifference between free circulating plasma and serum DNA in patients with colorectal liver metastasesAnticancer Res2002221A42142512017326
- MurtazaMDawsonSJTsuiDWNon-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNANature2013497744710811223563269
- DiehlFLiMDressmanDDetection and quantification of mutations in the plasma of patients with colorectal tumorsProc Natl Acad Sci USA200510245163681637316258065
- GiaconaMBRubenGCIczkowskiKARoosTBPorterDMSorensonGDCell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controlsPancreas199817189979667526
- SuzukiNKamatakiAYamakiJHommaYCharacterization of circulating DNA in healthy human plasmaClin Chim Acta20083871–2555817916343
- HoldenriederSStieberPChanLYCell-free DNA in serum and plasma: comparison of ELISA and quantitative PCRClin Chem20055181544154616040855
- HasselmannDORapplGTilgenWReinholdUExtracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serumClin Chem20014781488148911468248
- CherepanovaAVTamkovichSNBryzgunovaOEVlassovVVLaktionovPPDeoxyribonuclease activity and circulating DNA concentration in blood plasma of patients with prostate tumorsAnn N Y Acad Sci2008113721822118837950
- FrattiniMGallinoGSignoroniSQuantitative analysis of plasma DNA in colorectal cancer patients: a novel prognostic toolAnn N Y Acad Sci2006107518519017108210
- SozziGConteDLeonMQuantification of free circulating DNA as a diagnostic marker in lung cancerJ Clin Oncol200321213902390814507943
- SzpechcinskiAChorostowska-WynimkoJKupisWQuantitative analysis of free-circulating DNA in plasma of patients with resectable NSCLCExpert Opin Biol Ther201212 Suppl 1S3S922559166
- PaciMMaramottiSBellesiaECirculating plasma DNA as diagnostic biomarker in non-small cell lung cancerLung Cancer2009641929718804892
- CatarinoRFerreiraMMRodriguesHQuantification of free circulating tumor DNA as a diagnostic marker for breast cancerDNA Cell Biol200827841542118694299
- HashadDSorourAGhazalATalaatIFree circulating tumor DNA as a diagnostic marker for breast cancerJ Clin Lab Anal201226646747223143630
- SaiSIchikawaDTomitaHQuantification of plasma cell-free DNA in patients with gastric cancerAnticancer Res2007274C2747275117695442
- TomitaHIchikawaDIkomaDQuantification of circulating plasma DNA fragments as tumor markers in patients with esophageal cancerAnticancer Res2007274C2737274117695440
- ShapiroBChakrabartyMCohnEMLeonSADetermination of circulating DNA levels in patients with benign or malignant gastrointestinal diseaseCancer19835111211621206188527
- YoonKAParkSLeeSHKimJHLeeJSComparison of circulating plasma DNA levels between lung cancer patients and healthy controlsJ Mol Diagn200911318218519324991
- ZhangRShaoFWuXYingKValue of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: a meta-analysisLung Cancer201069222523120004997
- ChangHWLeeSMGoodmanSNAssessment of plasma DNA levels, allelic imbalance, and CA 125 as diagnostic tests for cancerJ Natl Cancer Inst200294221697170312441325
- AtamaniukJVidottoCTschanHBachlNStuhlmeierKMMüllerMMIncreased concentrations of cell-free plasma DNA after exhaustive exerciseClin Chem20045091668167015331502
- LaktionovPPTamkovichSNRykovaEYExtracellular circulating nucleic acids in human plasma in health and diseaseNucleosides Nucleotides Nucleic Acids2004236–787988315560076
- LoYMRainerTHChanLYHjelmNMCocksRAPlasma DNA as a prognostic marker in trauma patientsClin Chem200046331932310702517
- WuTLZhangDChiaJHTsaoKSunCFWuJTCell-free DNA: measurement in various carcinomas and establishment of normal reference rangeClin Chim Acta20023211–2778712031596
- LecomteTCezeNDorvalELaurent-PuigPCirculating free tumor DNA and colorectal cancerGastroenterol Clin Biol2010341266268120832215
- García-OlmoDCSamosJPicazoMGAsensioAITobosoIGarcía-OlmoDRelease of cell-free DNA into the bloodstream leads to high levels of non-tumor plasma DNA during tumor progression in ratsCancer Lett2008272113314018707808
- García-OlmoDCPicazoMGTobosoIAsensioAIGarcía-OlmoDQuantitation of cell-free DNA and RNA in plasma during tumor progression in ratsMol Cancer201312823374730
- AgostiniMPucciarelliSEnzoMVCirculating cell-free DNA: a promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapyAnn Surg Oncol20111892461246821416156
- FrattiniMGallinoGSignoroniSQuantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancerCancer Lett2008263217018118395974
- HoldenriederSStieberPvon PawelJCirculating nucleosomes predict the response to chemotherapy in patients with advanced non-small cell lung cancerClin Cancer Res20041018 Pt 15981598715447981
- HoldenriederSNagelDSchalhornAClinical relevance of circulating nucleosomes in cancerAnn N Y Acad Sci2008113718018918837945
- GormallyECabouxEVineisPHainautPCirculating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significanceMutat Res20076352–310511717257890
- WangBGHuangHYChenYCIncreased plasma DNA integrity in cancer patientsCancer Res200363143966396812873992
- UmetaniNKimJHiramatsuSIncreased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeatsClin Chem20065261062106916723681
- UmetaniNGiulianoAEHiramatsuSHPrediction of breast tumor progression by integrity of free circulating DNA in serumJ Clin Oncol200624264270427616963729
- ChenHSunLYZhengHQZhangQFJinXMTotal serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinomaPathology201244431832422531347
- JiangWWZahurakMGoldenbergDIncreased plasma DNA integrity index in head and neck cancer patientsInt J Cancer2006119112673267616991120
- PinzaniPSalviantiFZaccaraSCirculating cell-free DNA in plasma of melanoma patients: qualitative and quantitative considerationsClin Chim Acta201141223–242141214521839068
- GaoYJHeYJYangZLIncreased integrity of circulating cell-free DNA in plasma of patients with acute leukemiaClin Chem Lab Med201048111651165620831457
- ChanKCLeungSFYeungSWChanATLoYMPersistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patientsClin Cancer Res200814134141414518593992
- CasadioVCalistriDTebaldiMUrine Cell-Free DNA integrity as a marker for early bladder cancer diagnosis: Preliminary dataUrol Oncol2012 Epub119
- HoldenriederSBurgesAReichOSpelsbergFWStieberPDNA integrity in plasma and serum of patients with malignant and benign diseasesAnn N Y Acad Sci2008113716217018837942
- KopreskiMSBenkoFABorysDJKhanAMcGarrityTJGockeCDSomatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNAJ Natl Cancer Inst2000921191892310841827
- SorensonGDA review of studies on the detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancerAnn N Y Acad Sci2000906131610818588
- GarciaJMSilvaJMDominguezGSilvaJBonillaFHeterogeneous tumor clones as an explanation of discordance between plasma DNA and tumor DNA alterationsGenes Chromosomes Cancer200131330030111391802
- MaireFMicardSHammelPDifferential diagnosis between chronic pancreatitis and pancreatic cancer: value of the detection of KRAS2 mutations in circulating DNABr J Cancer200287555155412189555
- CastellsAPuigPMóraJK-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significanceJ Clin Oncol199917257858410080602
- GormallyEVineisPMatulloGTP53 and KRAS2 mutations in plasma DNA of healthy subjects and subsequent cancer occurrence: a prospective studyCancer Res200666136871687616818665
- ChenZFengJBuzinCHAnalysis of cancer mutation signatures in blood by a novel ultra-sensitive assay: monitoring of therapy or recurrence in non-metastatic breast cancerPLoS One200949e722019789704
- DiehlFSchmidtKChotiMACirculating mutant DNA to assess tumor dynamicsNat Med200814998599018670422
- LevyMBenesovaLLipskaLUtility of cell-free tumour DNA for post-surgical follow-up of colorectal cancer patientsAnticancer Res20123251621162622593440
- RyanBMLefortFMcManusRA prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow upGut200352110110812477769
- PageKHavaNWardBDetection of HER2 amplification in circulating free DNA in patients with breast cancerBr J Cancer201110481342134821427727
- DawsonSJTsuiDWMurtazaMAnalysis of circulating tumor DNA to monitor metastatic breast cancerN Engl J Med2013368131199120923484797
- GadgeelSMCoteMLSchwartzAGMatherlyLHWozniakABeplerGParameters for individualizing systemic therapy in non-small cell lung cancerDrug Resist Updat201013619620421051275
- RosellRVergnenegreALiuBBiomarkers in lung oncologyPulm Pharmacol Ther201023650851420471486
- MackPCHollandWSBurichRAEGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancerJ Thorac Oncol20094121466147219884861
- EstellerMSanchez-CespedesMRosellRSidranskyDBaylinSBHermanJGDetection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patientsCancer Res199959167709892187
- LoYMWongIHZhangJTeinMSNgMHHjelmNMQuantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reactionCancer Res199959163899390310463578
- CascianoIVinciADBanelliBCirculating tumor nucleic acids: perspective in breast CancerBreast care (Basel)201052758020847818
- RadpourRBarekatiZKohlerCHypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancerPLoS One201161e1608021283676
- WongIHLoYMZhangJDetection of aberrant p16 methylation in the plasma and serum of liver cancer patientsCancer Res199959171739892188
- SilvaJMDominguezGVillanuevaMJAberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patientsBr J Cancer19998081262126410376981
- TsutsuiMIizukaNMoribeTMethylated cyclin D2 gene circulating in the blood as a prognosis predictor of hepatocellular carcinomaClin Chim Acta20104117–851652020064498
- LingZQZhaoQZhouSLMaoWMMSH2 promoter hypermethylation in circulating tumor DNA is a valuable predictor of disease-free survival for patients with esophageal squamous cell carcinomaEur J Surg Oncol201238432633222265839
- GoesslCMüllerMStraubBMillerKDNA alterations in body fluids as molecular tumor markers for urological malignanciesEur Urol200241666867612074786
- IriyamaCTomitaAHoshinoHUsing peripheral blood circulating DNAs to detect CpG global methylation status and genetic mutations in patients with myelodysplastic syndromeBiochem Biophys Res Commun2012419466266922382018
- FieglHMillingerSMueller-HolznerECirculating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patientsCancer Res20056541141114515734995
- SchwarzenbachHMüllerVStahmannNPantelKDetection and characterization of circulating microsatellite-DNA in blood of patients with breast cancerAnn N Y Acad Sci20041022253215251935
- SunamiEShinozakiMHiganoCSMultimarker circulating DNA assay for assessing blood of prostate cancer patientsClin Chem200955355956719131636
- HamanaKUzawaKOgawaraKMonitoring of circulating tumour-associated DNA as a prognostic tool for oral squamous cell carcinomaBr J Cancer200592122181218415928666
- SchwarzenbachHMüllerVMilde-LangoschKSteinbachBPantelKEvaluation of cell-free tumour DNA and RNA in patients with breast cancer and benign breast diseaseMol Biosyst20117102848285421785770
- GarciaJMGarciaVSilvaJExtracellular tumor DNA in plasma and overall survival in breast cancer patientsGenes Chromosomes Cancer200645769270116607614
- TabackBO’DaySJBoasbergPDCirculating DNA microsatellites: molecular determinants of response to biochemotherapy in patients with metastatic melanomaJ Natl Cancer Inst200496215215614734706
- ChanKCLoYMCirculating tumour-derived nucleic acids in cancer patients: potential applications as tumour markersBr J Cancer200796568168517311021
- JonesKNourseJPKeaneCTumor-specific but not nonspecific cell-free circulating DNA can be used to monitor disease response in lymphomaAm J Hematol201287325826522213215
- LeiKIChanLYChanWYJohnsonPJLoYMQuantitative analysis of circulating cell-free Epstein-Barr virus (EBV) DNA levels in patients with EBV-associated lymphoid malignanciesBr J Haematol2000111123924611091207
- LoYMChanWYNgEKCirculating Epstein-Barr virus DNA in the serum of patients with gastric carcinomaClin Cancer Res2001771856185911448896
- LoYMPrognostic implication of pretreatment plasma/serum concentration of Epstein-Barr virus DNA in nasopharyngeal carcinomaBiomed Pharmacother200155736236511669497
- FlissMSUsadelHCaballeroOLFacile detection of mitochondrial DNA mutations in tumors and bodily fluidsScience200028754602017201910720328
- XiaPAnHXDangCXDecreased mitochondrial DNA content in blood samples of patients with stage I breast cancerBMC Cancer2009945420025731
- MithaniSKSmithIMZhouSMitochondrial resequencing arrays detect tumor-specific mutations in salivary rinses of patients with head and neck cancerClin Cancer Res200713247335734018094415
- JerónimoCNomotoSCaballeroOLMitochondrial mutations in early stage prostate cancer and bodily fluidsOncogene200120375195519811526508
- WarburgOWindFNegeleinEThe metabolism of tumors in the bodyJ Gen Physiol19278651953019872213
- ChiuRWChanLYLamNYQuantitative analysis of circulating mitochondrial DNA in plasmaClin Chem200349571972612709361
- LeeHCChangCMChiCWSomatic mutations of mitochondrial DNA in aging and cancer progressionAgeing Res Rev20109 Suppl 1S47S5820816876
- CopelandWCWachsmanJTJohnsonFMPentaJSMitochondrial DNA alterations in cancerCancer Invest200220455756912094550
- OkochiOHibiKUemuraTDetection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patientsClin Cancer Res2002892875287812231530
- KassaueiKHabbeNMullendoreMEKarikariCAMaitraAFeldmannGMitochondrial DNA mutations in pancreatic cancerInt J Gastrointest Cancer2006372–3576417827523
- MehraNPenningMMaasJvan DaalNGilesRHVoestEECirculating mitochondrial nucleic acids have prognostic value for survival in patients with advanced prostate cancerClin Cancer Res2007132 Pt 142142617255261
- HibiKNakayamaHYamazakiTMitochondrial DNA alteration in esophageal cancerInt J Cancer200192331932111291064
- HibiKNakayamaHYamazakiTDetection of mitochondrial DNA alterations in primary tumors and corresponding serum of colorectal cancer patientsInt J Cancer200194342943111745425
- KohlerCRadpourRBarekatiZLevels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumorsMol Cancer2009810519922604
- ZachariahRRSchmidSBuerkiNRadpourRHolzgreveWZhongXLevels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumorsObstet Gynecol2008112484385018827127
- EllingerJMüllerDCMüllerSCCirculating mitochondrial DNA in serum: a universal diagnostic biomarker for patients with urological malignanciesUrol Oncol201230450951520870429
- WieczorekAJSitaramamVMachleidtWRhynerKPerruchoudAPBlockLHDiagnostic and prognostic value of RNA-proteolipid in sera of patients with malignant disorders following therapy: first clinical evaluation of a novel tumor markerCancer Res19874723640764122445471
- LässerCAlikhaniVSEkströmKHuman saliva, plasma and breast milk exosomes contain RNA: uptake by macrophagesJ Transl Med20119921235781
- KopreskiMSBenkoFAKwakLWGockeCDDetection of tumor messenger RNA in the serum of patients with malignant melanomaClin Cancer Res1999581961196510473072
- LoKWLoYMLeungSFAnalysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinomaClin Chem1999458 Pt 11292129410430801
- DasíFLledóSGarcía-GraneroEReal-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: a simple blood test to monitor disease in cancer patientsLab Invest200181576776911351048
- SilvaJMRodriguezRGarciaJMDetection of epithelial tumour RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cellsGut200250453053411889075
- LledóSMGarcia-GraneroEDasíFReal time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA in patients with colorectal cancerColorectal Dis20046423624215206965
- SilvaJMDominguezGSilvaJDetection of epithelial messenger RNA in the plasma of breast cancer patients is associated with poor prognosis tumor characteristicsClin Cancer Res2001792821282511555599
- ChenXQBonnefoiHPelteMFTelomerase RNA as a detection marker in the serum of breast cancer patientsClin Cancer Res20006103823382611051224
- DasíFMartínez-RodesPMarchJAReal-time quantification of human telomerase reverse transcriptase mRNA in the plasma of patients with prostate cancerAnn N Y Acad Sci2006107520421017108213
- March-VillalbaJAMartínez-JabaloyasJMHerreroMJSantamaríaJAliñoSFDasíFPlasma hTERT mRNA discriminates between clinically localized and locally advanced disease and is a predictor of recurrence in prostate cancer patientsExpert Opin Biol Ther201212 Suppl 1S69S7722559196
- KopreskiMSBenkoFAGockeCDCirculating RNA as a tumor marker: detection of 5T4 mRNA in breast and lung cancer patient serumAnn N Y Acad Sci200194517217811708475
- ChinnappaPTagubaLArciagaRDetection of thyrotropin-receptor messenger ribonucleic acid (mRNA) and thyroglobulin mRNA transcripts in peripheral blood of patients with thyroid disease: sensitive and specific markers for thyroid cancerJ Clin Endocrinol Metab20048983705370915292293
- ReddiKKHollandJFElevated serum ribonuclease in patients with pancreatic cancerProc Natl Acad Sci USA1976737230823101065880
- TsuiNBNgEKLoYMStability of endogenous and added RNA in blood specimens, serum, and plasmaClin Chem200248101647165312324479
- NgEKTsuiNBLamNYPresence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individualsClin Chem20024881212121712142376
- TzimagiorgisGMichailidouEZKritisAMarkopoulosAKKouidouSRecovering circulating extracellular or cell-free RNA from bodily fluidsCancer Epidemiol201135658058921514265
- TerrinLRampazzoEPucciarelliSRelationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic diseaseClin Cancer Res200814227444745119010861
- MiuraNNakamuraHSatoRClinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor marker for lung cancerCancer Sci200697121366137317052260
- MiuraNMaruyamaSOyamaKDevelopment of a novel assay to quantify serum human telomerase reverse transcriptase messenger RNA and its significance as a tumor marker for hepatocellular carcinomaOncology200772 Suppl 1455118087181
- PucciarelliSRampazzoEBriaravaMTelomere-specific reverse transcriptase (hTERT) and cell-free RNA in plasma as predictors of pathologic tumor response in rectal cancer patients receiving neoadjuvant chemoradiotherapyAnn Surg Oncol20121993089309622395986
- El-HefnawyTRajaSKellyLCharacterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnosticsClin Chem200450356457314718398
- El-AbdEEl-TahanRFahmyLSerum metastasin mRNA is an important survival predictor in breast cancerBr J Biomed Sci2008652909419055112
- FleischhackerMBeinertTErmitschMDetection of amplifiable messenger RNA in the serum of patients with lung cancerAnn N Y Acad Sci200194517918811708476
- HonmaHKandaTItoHSquamous cell carcinoma-antigen messenger RNA level in peripheral blood predicts recurrence after resection in patients with esophageal squamous cell carcinomaSurgery2006139567868516701102
- DongZZYaoDFYaoMClinical impact of plasma TGF-beta1 and circulating TGF-beta1 mRNA in diagnosis of hepatocellular carcinomaHepatobiliary Pancreat Dis Int20087328829518522884
- IorioMVCroceCMMicroRNAs in cancer: small molecules with a huge impactJ Clin Oncol200927345848585619884536
- ChenXBaYMaLCharacterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseasesCell Res20081810997100618766170
- NgEKChongWWJinHDifferential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screeningGut200958101375138119201770
- HuangZHuangDNiSPengZShengWDuXPlasma microRNAs are promising novel biomarkers for early detection of colorectal cancerInt J Cancer2010127111812619876917
- KosakaNIguchiHOchiyaTCirculating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosisCancer Sci2010101102087209220624164
- HuZChenXZhaoYSerum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancerJ Clin Oncol201028101721172620194856
- WangZXBianHBWangJRChengZXWangKMDeWPrognostic significance of serum miRNA-21 expression in human non-small cell lung cancerJ Surg Oncol2011104784785121721011
- LiuXGZhuWYHuangYYHigh expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancerMed Oncol201229261862621516486
- RothCStückrathIPantelKIzbickiJRTachezyMSchwarzenbachHLow levels of cell-free circulating miR-361-3p and miR-625* as blood-based markers for discriminating malignant from benign lung tumorsPLoS One201276e3824822675530
- WangFZhengZGuoJDingXCorrelation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumorGynecol Oncol2010119358659320801493
- MitchellPSParkinRKKrohEMCirculating microRNAs as stable blood-based markers for cancer detectionProc Natl Acad Sci USA200810530105131051818663219
- BandresEBitarteNAriasFmicroRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cellsClin Cancer Res20091572281229019318487
- YangHGuJWangKKMicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinomaClin Cancer Res200915185744575219737949
- LawrieCHGalSDunlopHMDetection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphomaBr J Haematol2008141567267518318758
- ZandbergaEKozirovskisVĀbolsAAndrējevaDPurkalneGLinēACell-free microRNAs as diagnostic, prognostic, and predictive biomarkers for lung cancerGenes Chromosomes Cancer201352435636923404859
- LuJGetzGMiskaEAMicroRNA expression profiles classify human cancersNature2005435704383483815944708
- RosenfeldNAharonovRMeiriEMicroRNAs accurately identify cancer tissue originNat Biotechnol200826446246918362881
- WeberJABaxterDHZhangSThe microRNA spectrum in 12 body fluidsClin Chem201056111733174120847327
- XieLQianXLiuBMicroRNAs: novel biomarkers for gastrointestinal carcinomasMol Cell Biochem20103411–229129920422260
- MoMHChenLFuYWangWFuSWCell-free Circulating miRNA Biomarkers in CancerJ Cancer2012343244823074383
- TokarzPBlasiakJThe role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatmentActa Biochim Pol201259446747423173124
- MattocksCJMorrisMAMatthijsGA standardized framework for the validation and verification of clinical molecular genetic testsEur J Hum Genet201018121276128820664632
- García-OlmoDGarcía-OlmoDCOntañónJMartinezEVallejoMTumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasisHistol Histopathol19991441159116410506932
- García-OlmoDCDomínguezCGarcía-ArranzMCell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cellsCancer Res201070256056720068178
- García-OlmoDGarcía-OlmoDCDomínguez-BerzosaCGuadalajaraHVegaLGarcía-ArranzMOncogenic transformation induced by cell-free nucleic acids circulating in plasma (genometastasis) remains after the surgical resection of the primary tumor: a pilot studyExpert Opin Biol Ther201212 Suppl 1S61S6822568822
- SilvaJGarciaVRodriguezMAnalysis of exosome release and its prognostic value in human colorectal cancerGenes Chromosomes Cancer201251440941822420032
- MittelbrunnMGutiérrez-VázquezCVillarroya-BeltriCUnidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cellsNat Commun2011228221505438
- PeinadoHAlečkovićMLavotshkinSMelanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through METNat Med201218688389122635005