Abstract
Short-rib polydactyly syndromes (SRPS) arise from mutations in genes involved in retrograde intraflagellar transport (IFT) and basal body homeostasis, which are critical for cilia assembly and function. Recently, mutations in WDR34 or WDR60 (candidate dynein intermediate chains) were identified in SRPS. We have identified and characterized Tctex1d2, which associates with Wdr34, Wdr60 and other dynein complex 1 and 2 subunits. Tctex1d2 and Wdr60 localize to the base of the cilium and their depletion causes defects in ciliogenesis. We propose that Tctex1d2 is a novel dynein light chain important for trafficking to the cilium and potentially retrograde IFT and is a new molecular link to understanding SRPS pathology.
Keywords:
Introduction
Primary cilia are microtubule-based organelles that project from almost all vertebrate cells into the extracellular matrix and are critical for sensing the external environment (including photosensation and mechanosensation) and transducing these signals into cellular decisions (including cell growth and differentiation).Citation1 Of interest, mutation of the machinery critical for the establishment and function of the cilia manifests itself as a group of disorders collectively known as ciliopathies.Citation2 Among the ciliopathy disease spectrum are the short-rib polydactyly syndromes (SRPS) that include Jeune asphyxiating thoracic dystrophy (JATD) and Ellis–van Creveld syndrome (EVC), which are characterized by shortened ribs and tubular bones, a constricted thoracic cage, and polydactyly.Citation3
To date at least 10 genes have been identified to be responsible for SRPS, most of which are involved in retrograde intraflagellar transport (IFT) (IFT43, IFT122, WDR19, WDR35, TTC21B, and DYNC2H1) or basal body homeostasis (NEK1, EVC, and EVC2).Citation3 Recently, mutations in the WDR34 and WDR60 genes were identified in SRPS.Citation4,5 Both Wdr34 and Wdr60 localize to the base of the cilium in human ciliated cells and WDR60 mutant cells from SRPS affected individuals have a drastic decrease in their ability to form cilia.Citation4,5 Although, there is limited molecular characterization of Wdr34 and Wdr60 in mammals, the Chlamydomonas reinhardtii orthologs of Wdr34 (FAP133) and Wdr60 (FAP163) have been characterized as potential dynein intermediate chains required for retrograde IFT.Citation6,7
Within the context of ciliogenesis, cytoplasmic dynein 1 complex (Dync1) is less studied than cytoplasmic dynein 2 complex (Dync2), which is involved in retrograde IFT. However, there is growing evidence indicating that several components are shared between Dync1 and Dync2 complexes including the light chains Tctex1 and Dynll1 and the intermediate chain Wdr34.Citation5,8,9 For example, Tctex1 and Dynll1 have been implicated in regulating cilium length, where depletion of Tctex1 leads to elongated cilia and depletion of Dynll1 leads to a decrease in ciliation.Citation9,10 Thus, highlighting the growing consensus that light chains have multiple functions in trafficking within the cell and the cilium through their interactions with Dync1 and Dync2 complexes.
Recently, a new class of candidate light chains that contain a conserved domain similar to the C-terminus of Tctex1 have been annotated, which include Tctex1d1-4 (Tctex1 domain containing 1-4). However, with the exception of Tctex1d4s characterization as a protein phosphatase 1 interacting proteinCitation11,12, there has been no known molecular characterization of this protein family. Here, we define a function for Tctex1d2 in ciliogenesis. Tctex1d2 associates with Wdr34, Wdr60, and other subunits of Dync1 and Dync2 and colocalizes with Wdr60 to microtubule organizing centers during interphase, the mitotic spindle poles during cell division, and the base of the cilium in ciliated cells. Interestingly, depletion of Tctex1d2 and Wdr60 leads to defective cilia formation. Additionally, the proper localization of Tctex1d2 to the base of the cilium depends on microtubules and Wdr60. We propose a model where Tctex1d2 is a Dync1 and Dync2 light chain, which functions as a substrate adaptor for transporting cargo to the cilium and potentially within the cilium that is thus essential for proper ciliogenesis. Therefore, Tctex1d2 represents a novel molecular link that couples the cellular motor transport machinery to ciliopathies like SRPS.
Results
Tctex1d2 associates with Wdr34, Wdr60, and cytoplasmic dynein complex 1 and 2
Our proteomic studies aimed at identifying novel microtubule associated proteins led us to discover MGC33212, a hypothetical uncharacterized protein with a Tctex1 domain at its C-terminus.Citation13 Tctex1 (known as Dynlt1) is a well-characterized dynein light chain that utilizes its C-terminal Tctex1 domain to bind dynein intermediate chains and its N-terminal domain to bind specific cargo, which has important roles in cytoplasmic trafficking and cilia formation.Citation9,14-16 The human genome encodes 4 Tctex1 domain-containing proteins: Tctex1d1–4, MGC33212 is also referred to as Tctex1d2 (). Tctex1d2 shares 20% amino acid identity with Tctex1 (Fig. S1). Tctex1d2 also shares 29%, 23%, and 29% identity with Tctex1d1, Tctex1d3, and Tctex1d4 respectively (Fig. S1). Although previous bioinformatic, genomic, and proteomic studies aimed at defining the ciliome had implicated Tctex1d2 in ciliation, it remained completely uncharacterized.Citation17-22
Figure 1 (See previous page). Tctex1d2 and Wdr60 associate with cytoplasmic dynein complex 1 and 2. (A) Schematic of Tctex1 domain containing proteins. All members have a carboxyl terminal Tctex1 domain and a variable N- terminal domain implicated in cargo binding. The number of amino acid residues is indicated for each protein. (B) LAP-Tctex1d2 tandem affinity purification. MW= molecular weight marker, HSS= high spin supernatant, E= final eluates. Asterisk denotes Tctex1d2. (C) Cytoscape analysis showing that Tctex1d2 interacting proteins can be grouped into 6 major categories: tubulin isoforms, microtubule binding proteins, centrosome associated proteins, proteins associated with ciliogenesis, cytoplasmic dynein complex 1 and 2 (Dync1 and Dync2) subunits, and proteins implicated in other cellular roles. (D) Summary of mass spectrometry data for Dync1 and Dync2 components identified in the LAP-Tctex1d2 purification, including protein name, number of peptides identified, number of unique peptides, and the percent protein coverage. (E) HEK293 or HEK293 LAP-Tctex1d2 expressing cell extracts were used to perform immunoprecipitations (IPs) with anti-GFP antibodies. IPs were resolved by SDS PAGE, transferred to a PVDF membrane, and immunoblotted with indicated antibodies. (F and G) Reciprocal co-IPs of endogenous Tctex1d2 and Wdr60 from HEK293 cells, using anti-Tctex1d2 or anti-Wdr60 antibodies. Western blot analysis shows that Tctex1d2 and Wdr60 co-IP with each other and both IP Dync1 and Dync2 subunits. See also Fig. S1 and Table S1. (H) In vitro binding assays testing the binding of in vitro transcribed/translated 35S-radiolabeled HA-tagged full-length Tctex1d2, N-terminal Tctex1d2 (amino acids 1-40), and C-terminal Tctex1d2 (amino acids 41-142) with FLAG-tagged full-length Wdr34 and Wdr60. Binding was detected by radiometric analyses. I indicates Tctex1d2 input only, numbers 1-3 indicate incubation with Wdr60 (1), Wdr34 (2), or Luciferase (3).
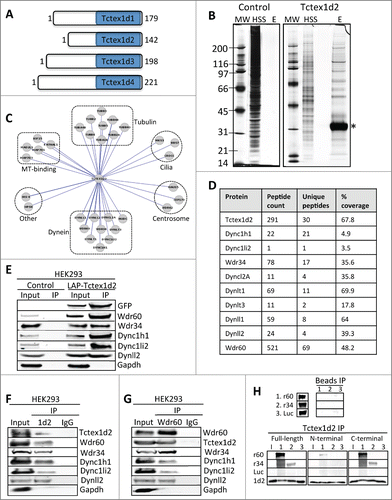
To define the cellular role of Tctex1d2, we began by analyzing its protein-protein interactions. To do this, we generated a doxycycline-inducible localization and affinity purification (LAP= EGFP-TEV-S-Peptide)-tagged-Tctex1d2 HEK293 stable cell line.Citation23 The LAP-Tctex1d2 HEK293 stable cell line was used to express and tandem affinity purify LAP-Tctex1d2 (first affinity purification using anti-GFP antibody conjugated beads and the second affinity purification using S-protein conjugated beads). Eluates from these purifications were separated by gel chromatography and 10 gel slices corresponding to the entire eluates were excised and analyzed by mass spectrometry (). Visualization of the proteins we identified as Tctex1d2 copurifying proteins using CytoscapeCitation24 showed that Tctex1d2 interacting proteins could be grouped into 6 major categories: tubulin isoforms, microtubule binding proteins, centrosome associated proteins, ciliogenesis associated proteins, Dync1 and Dync2 subunits, and proteins with other cellular roles ( and Table S1). This analysis identified Tctex1d2 as one of the most abundant proteins in this purification (291 total peptides, 30 unique peptides covering 67.8% of Tctex1d2) (). Tctex1d2 associated with Dync1 and Dync2 subunits including heavy chains (Dync1h1 and Dync2h1), intermediate chains (Wdr34 and Wdr60), light intermediate chain (Dync1li2), and light chains (Dynll1, Dynll2, Dynlrb1, Dynlt1, and Dynlt3) ( and Table S1). Interestingly, Tctex1d2 also associated with proteins critical for ciliogenesis, most of which mutation/dysfunction is associated with ciliopathies, inclduding MKS3 (Meckel–Gruber syndrome 3), BBS7 (Bardet–Biedl syndrome 7), Wdr34, Wdr60 and CROCC (ciliary rootlet coiled coil) ( and Table S1).Citation4,5,25-27
To further validate Tctex1d2s interaction with Wdr34, Wdr60, and other Dync1 and Dync2 subunits, we performed LAP(EGFP-TEV-S-Peptide)-Tctex1d2 immunoprecipitations (IPs) using anti-GFP antibody conjugated beads and immunoblotted for Wdr34, Wdr60 and dynein components. Indeed, Wdr34, Wdr60, Dync1h1, Dync1li2, and Dynll2 co-IPd with LAP-Tctex1d2 in anti-GFP IPs (). To determine if these interactions were observed with endogenous proteins, we performed reciprocal co-IPs with anti-Tctex1d2 and anti-Wdr60 antibodies. Consistently, Tctex1d2 IPd Wdr60, Wdr34, and other Dync1 and Dync2 components and Wdr60 IPd Tctex1d2, Wdr34, and other Dync1 and Dync2 components ( and ). Next, we asked if Tctex1d2 bound directly to Wdr34 and Wdr60 and what region of Tctex1d2 was required for this interaction. To do this, we generated full-length and N-terminal (amino acids 1–40) and C-terminal (amino acids 41–142, contains Tctex1 domain) Tctex1d2 truncation mutants and analyzed their binding to Wdr34 and Wdr60 in vitro. Interestingly, only the full-length Tctex1d2 and its C-terminal fragment bound to both Wdr34 and Wdr60 (). Whereas, the N-terminal Tctex1d2 fragment only minimally bound to Wdr60 (). These data indicated that similar to other Tctex1 domain containing proteins, Tctex1d2 bound to Wdr34 and Wdr60 via its C-terminal Tctex1 domain.
Tctex1d2 and Wdr60 localize to the MTOC and spindle poles in non-ciliated cells
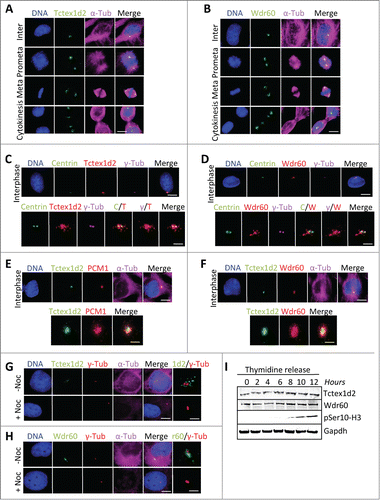
To further understand the cellular role of Tctex1d2, we focused on its association with the poorly characterized Wdr60 protein (561 Wdr60 peptides identified by mass spectrometry, ) and analyzed their subcellular localization throughout the cell cycle. Immunofluorescence microscopy of HeLa cells with anti-Tctex1d2 or anti-Wdr60 antibodies indicated that both proteins localized to the MTOC in interphase, the spindle poles during mitosis, and the centrosomes and cytokinetic bridge microtubules during cytokinesis ( and ). Consistently, overexpressed (stable or transient) LAP(EGFP-TEV-S-Peptide)-Tctex1d2 also showed a similar localization pattern (Fig. S2). To further define the Tctex1d2 and Wdr60 subcellular localization, we performed colocalization studies with centrosome markers ( and ). Tctex1d2 and Wdr60 localized as puncta surrounding centrin and gamma tubulin, which stained the centrioles ( and ). Furthermore, Tctex1d2 co-localized with PCM1, indicating that it may associate with pericentriolar satellites (). Consistent with Tctex1d2s interaction with Wdr60, LAP-Tctex1d2 co-localized with Wdr60 at the MTOC (). Since Tctex1d2 was associating with microtubule-dependent minus end directed motors (Dync1 and Dync2), we asked if the localization of Tctex1d2 and Wdr60 to the MTOC was dependent on microtubules. Indeed, Tctex1d2 and Wdr60 were absent from the MTOC in cells treated with the microtubule depolymerizing agent nocodazole ( and ). Although Tctex1d2 and Wdr60 had a dynamic cell cycle-dependent localization, their protein levels remained constant throughout the cell cycle with no obvious signs of modifications (). Together these results indicated that Tctex1d2 and Wdr60 were stable throughout the cell cycle, associated with dynein, and localized to the MTOC in a microtubule-dependent manner.
Figure 2 (See previous page). Tctex1d2 and Wdr60 localize to microtubule organizing centers. (A and B) Immunofluorescence microscopy of fixed HeLa cells stained with Hoechst 33342 DNA dye, and anti-α-tubulin, anti-Tctex1d2 (A), or anti-Wdr60 (B) antibodies. Images show the cell cycle subcellular localization of Tctex1d2 (A) and Wdr60 (B). Note that both proteins localize to the microtubule-organizing center (MTOC) in interphase, the spindle poles during mitosis (prometaphase, metaphase) and cytokinesis, and near the cytokinetic bridge during cytokinesis. Bar= 5 μm. (C and D) Immunofluorescence microscopy of fixed interphase HeLa cells stained with Hoechst 33342 DNA dye, anti-γ-tubulin, anti-Centrin, anti-Tctex1d2 (C), or anti-Wdr60 (D) antibodies. Bar= 5 μm. Lower panels show a zoom view of the MTOC region. Bar= 2 μm. (E and F) Immunofluorescence microscopy of fixed interphase HEK293 cells expressing LAP-Tctex1d2 fixed and stained with Hoechst 33342 DNA dye, anti-α-tubulin, anti-PCM1 (E), or anti-Wdr60 (F) antibodies. Bar= 5 μm. Lower panels show a zoom view of the MTOC region. Bar= 2 μm. (G and H) Immunofluorescence microscopy of fixed interphase HeLa cells treated with or without Nocodazole, fixed, and stained with Hoechst 33342 DNA dye, anti-γ-tubulin, anti-α-tubulin, anti-Tctex1d2 (G), or anti-Wdr60 (H) antibodies. Bar= 5μm. Rightmost panels show a zoom view of the MTOC region. Bar= 2 μm. (I) Analysis of Tctex1d2 and Wdr60 protein levels throughout the cell cycle. HeLa cells were synchronized in G1/S, released into the cell cycle and cells were harvested at the indicated time points. Protein extracts were prepared, resolved by SDS PAGE, transferred to a PVDF membrane and immunoblotted with indicated antibodies. Note that Tctex1d2 and Wdr60 protein levels remain constant throughout the cell cycle. See also Fig. S2.
In ciliated cells Tctex1d2 associates with Wdr34, Wdr60 and cytoplasmic dynein complex 2 and localizes to the base of the cilium
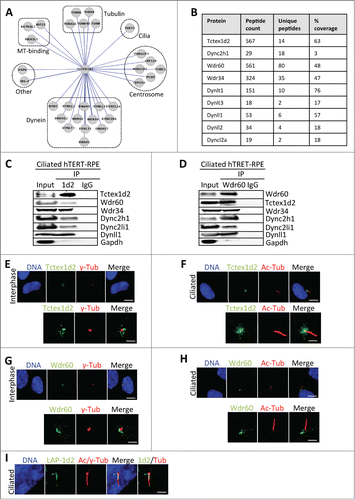
Because of Tctex1d2s potential connection to ciliogenesis and its interaction with the Wdr60 and Wdr34 proteins that are found mutated in ciliopathies like SRPS,Citation4,17-22 we asked if Tctex1d2 associated with the ciliary Dync2 complex in ciliated cells. To do this, we performed a tandem affinity purification of LAP(EGFP-TEV-S-Peptide)-Tctex1d2 from ciliated cells, analyzed the Tctex1d2 interactors by mass spectrometry, and visualized the Tctex1d2 interactome via CytoscapeCitation24 ( and and Table S1). This analysis revealed that Tctex1d2 associated with ciliary Dync2 complex subunits, including Dync2h1, Wdr60, Wdr34, and Dynll1 ( and and Table S1). This was verified by performing reciprocal co-IPs with anti-Tctex1d2 and anti-Wdr60 antibodies from ciliated hTERT-RPE cells, where Tctex1d2 IPd Dync2h1, Wdr60, Wdr34, Dync2li1 and Dynll1, and Wdr60 IPd Tctex1d2, Dync2h1, Wdr34, Dync2li1 and Dynll1 ( and ). Next, we analyzed the subcellular localization of Tctex1d2 and Wdr60 in non-ciliated and ciliated hTERT-RPE cells. Both Tctex1d2 and Wdr60 localized to the region surrounding the centrioles in interphase cells and the centrioles/basal body in ciliated cells ( and ). Although we were unable to detect Tctex1d2 within the cilium with anti-Tctex1d2 antibodies, overexpression of LAP(EGFP-TEV-S-Peptide)-Tctex1d2 led to the localization of LAP-Tctex1d2 to the ciliary axoneme (), similar to what had been observed with Wdr60 overexpression.Citation4 These results indicated that Tctex1d2 associated with the ciliary Dync2 complex in ciliated cells and that it localized to the base of the cilium and possibly within the cilium.
Figure 3. Tctex1d2 and Wdr60 associate with cytoplasmic dynein complex 2 and localize to the base of the cilia in ciliated cells. (A) Cytoscape analysis showing that Tctex1d2 interacts with the Dync2 complex along with tubulin isoforms, microtubule binding proteins, centrosome associated proteins, proteins associated with ciliogenesis, and proteins implicated in other cellular roles in ciliated cells. (B) Summary of mass spectrometry data for Dync2 subunits identified in the LAP-Tctex1d2 purification from ciliated HEK293 cells, including protein name, number of peptides identified, number of unique peptides, and percent protein coverage. (C and D) Reciprocal co-IPs of endogenous Tctex1d2 and Wdr60 from ciliated hTERT-RPE cells, using anti-Tctex1d2 or anti-Wdr60 antibodies. Immunoblot analysis shows that Tctex1d2 and Wdr60 co-IP with each other and both IP Dync2 subunits. (E–H) Immunofluorescence microscopy of fixed interphase or ciliated hTERT-RPE cells stained with Hoechst 33342 DNA dye, anti-γ-tubulin, anti-Tctex1d2 (E and F), or anti-Wdr60 (G and H) antibodies. Bar= 5 μm. Lower panels show a zoom view of the MTOC region (E and G) or the base of the cilium (F and H). Bar= 2 μm. (I) Immunofluorescence microscopy of fixed ciliated hTERT-RPE cells expressing LAP-Tctex1d2 showing that Tctex1d2 localizes to the ciliary base and axoneme. Cell staining was as described in (C). Bar= 2 μm.
Tctex1d2 and Wdr60 are required for ciliogenesis
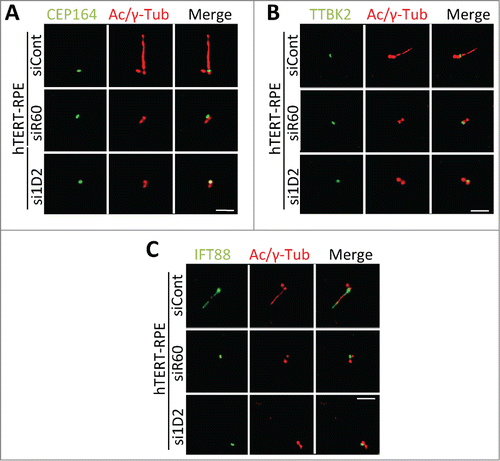
Next, we asked if Tctex1d2 or Wdr60 were required for cilia formation by depleting Tctex1d2 or Wdr60 and monitoring ciliation in hTERT-RPE cells. First, we identified siRNA oligonucleotides which reduced Tctex1d2 or Wdr60 protein levels to less than 10% compared to control non-targeting siRNA, oligonucleotides si1D2 and siR60 respectively ( and and Fig. S3). Interestingly, depletion of either Tctex1d2 or Wdr60 led a significant reduction in the percentage of ciliated cells (siID2= 27.3±8.5, p=.003 and siR60= 21±7.0, p=.001 compared to siControl= 64±5.0) ( and ). However, depletion of both Tctex1d2 and Wdr60 did not lead to a significant reduction in ciliation compared to depletion of Wdr60 alone (). Also, whereas depletion of Tctex1d2 had no effect on the localization of Wdr60 (localized to a region adjacent to the basal body), depletion of Wdr60 led to a reduced localization of Tctex1d2 to the basal body ( and ). Together, these results indicated that Tctex1d2 and Wdr60 were required for cilia formation and that the proper localization of Tctex1d2 to the base of the cilium was dependent on Wdr60. Finally, we asked if depletion of Tctex1d2 or Wdr60 affected the recruitment of factors necessary for the initiation of ciliogenesis to the mother centriole, including CEP164 and TTBK2.Citation28,29 However, CEP164 and TTBK2 were localized to the mother centriole in cells depleted for Tctex1d2 or Wdr60 ( and ). Additionally, in control cells IFT88 localized to the mother centriole and within the cilium (as previously shownCitation30), whereas in Tctex1d2 or Wdr60 depleted cells it only localized to the mother centriole, as no cilium was formed in these cells (). Based on these markers of early ciliogenesis it appears that Tctex1d2 and Wdr60 are not necessary for the early steps of ciliogenesis, albeit an extensive analysis with additional cilia markers would be necessary to fully support this idea.
Figure 4. Depletion of Tctex1d2 or Wdr60 leads to ciliation defects. (A and B) siRNA knockdown of Tctex1d2 or Wdr60 protein levels. Immunoblot analysis showing that siRNA pool oligonucleotides targeting TCTEX1D2 (si1D2) or WDR60 (siR60) deplete Tctex1d2 or Wdr60 protein levels in hTERT-RPE cells compared to non-targeting control siRNA (siCont). (C and D) Immunofluorescence of ciliated hTERT-RPE cells treated with siCont, si1D2, or siR60 for 48 hours, starved for 24 hours, and stained with Hoechst 33342 DNA dye, anti-γ-tubulin, anti-acetylated (Ac)-tubulin, and anti-Tctex1d2 (C) or anti-Wdr60 (D). Note the lack of Tctex1d2 at the base of the cilium in si1D2 treated cells and defective cilia formation. Similarly the lack of Wdr60 at the base of the cilium in siR60 treated cells and defective cilia formation. Scale bar = 5 μm. Rightmost panels show a zoom view of the base of the cilium. Bar= 2 μm. (E) Quantitation of the percentage of ciliated cells siCont, si1D2, siR60 and si1D2/siR60-treated cells. Data represent the average ± SD of 3 independent experiments, 100 cells counted for each, p values are as indicated. NS indicates not statistically significant. (F and G) Immunofluorescence of ciliated hTERT-RPE cells treated with siCont, si1D2, or siR60 for 48 hours, starved for 24 hours, and stained with Hoechst 33342 DNA dye, anti-γ-tubulin, anti-acetylated (Ac)-tubulin, and anti-Wdr60 (F) or anti-Tctex1d2 (G). Note Wdr60 localizes to the base of the cilium in si1D2 treated cells, whereas Tctex1d2 is mostly absent from the base of the cilium in siR60 treated cells. Scale bar = 5 μm. Rightmost panels show a zoom view of the base of the cilium. Bar= 2 μm.
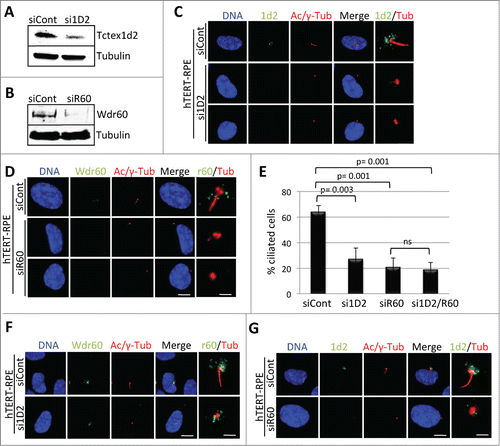
Figure 5. Depletion of Tctex1d2 or Wdr60 does not affect the recruitment of some factors necessary for the early steps of ciliogenesis. (A–C) Immunofluorescence microscopy of ciliated hTERT-RPE cells treated with siCont, si1D2, or siR60 for 48 hours, starved for 24 hours, and stained with Hoechst 33342 DNA dye, anti-γ-tubulin, anti-acetylated (Ac)-tubulin, and anti-CEP164 (A), anti-TTBK2 (B), or anti-IFT88 (C). Note that all proteins localize to the mother centriole in si1D2 and siR60 treated cells. Scale bar = 5 μm.
Discussion
The molecular mechanisms underlying many ciliopathies remain to be determined. Our data indicates that Tctex1d2 associates with SRPS proteins Wdr60, Wdr34, and Dync2h1, all of which have been implicated in retrograde IFT and whose mutation causes ciliopathies including Short-rib polydactyly syndrome (SRPS).Citation4,5,31 Tctex1d2 also associated with other subunits of the Dync2 complex and its localization to the base of the cilium was microtubule and Wdr60-dependent. Finally, both Tctex1d2 and Wdr60 were found to be important for ciliogenesis. We favor a model where Tctex1d2 functions as a cargo adaptor that interacts with Wdr60/Wdr34 containing Dync1/Dync2 to deliver specific cargo necessary for ciliogenesis to the base of the cilium and potentially has a role in IFT within the cilium. Although, we were unable to detect endogenous Tctex1d2 or Wdr60 within the cilium, a previous study showed that overexpressed Wdr60 localized to the ciliary axoneme.Citation4 Similarly, overexpressed Tctex1d2 localized to the base of the cilium and ciliary axoneme. Thus, it is possible that these proteins could associate with the retrograde IFT machinery in low abundance and that these interactions could be critical for ciliogenesis.
The cargoes transported by Tctex1d2 that are important for ciliogenesis remain to be elucidated. However, MKS3, BBS7, and CROCC are potential candidates as all have critical functions in ciliogenesis and were found to associate with Tctex1d2 in biochemical purifications.Citation4,5,25,26 Mutation of proteins involved in centrosomal function have also been linked to SRPSCitation3, thus Tctex1d2-interacting and centrosomal associated proteins like Wdr62, Cep170, Haus5, CEP128, CEP250, and Pericentrin could also be potential cargoes. However, the localization of CEP164, TTBK2 and IFT88 to the mother centriole in Tctex1d2 and Wdr60 depleted cells would indicate that the centrosomes have matured and are competent for ciliogenesis. Thus, an in depth analysis of ciliogenesis markers will be necessary to fully determine the step at which Tctex1d2 and Wdr60 are necessary during ciliogenesis. Interestingly, in cycling cells both Tctex1d2 and Wdr60 associated with Dync1 subunits and localized to MTOCs, spindle poles and intracellular bridge microtubules, thus Tctex1d2 and Wdr60 are likely to have roles in intracellular trafficking independent of their involvement in ciliogenesis that should also be explored further.
Materials and Methods
Cell culture
hTERT-RPE, HeLa and HEK293 cell were grown in F12:DMEM 50:50 (GIBCO) with 10% FBS, 2mM L-glutamine and antibiotics in 5% CO2 at 37°C. Cells were synchronized in G1/S by treatment with 2 mM thymidine (Sigma-Aldrich) for 18-hours. For microtubule depolymerization cells were treated with 300 nM Nocodazole (Sigma). For siRNA treatments, Dharmacon ON-TARGETplus siRNA smart pools and individual oligos: D-001810-10 (control non-targeting siRNA); L-015945-01, J-015945-9, J-015945-10, J-015945-11, J-015945-12 (siRNA targeting TCTEX1D2); and L-016701-02, J-016701-17, J-016701-18, J-016701-19, J-016701-20 (siRNA targeting WDR60) were used as previously described by Torres et al.Citation32
Generation of stable cell lines
HEK293 LAP(EGFP-TEV-S-Peptide)-Tctex1d2 stable cell lines were generated according to Torres et al.Citation23 Full-length TCTEX1D2 (amino acid residues 1-142) was fused to the C-terminus of the S-Peptide to generate the pGLAP1-TCTEX1D2 vector that was used to establish the T-REx LAP-TCTEX1D2 stable cell line.Citation23
Immunoprecipitations, LAP purifications and LC-MS/MS analyses
For biochemical purifications from ciliated cells, ciliation was followed in parallel by immunofluorescence microscopy prior to preparation of cell extracts. For both ciliated and non-ciliated cells, whole cell extracts were prepared in LAP300 lysis buffer (50 mM Hepes pH 7.4, 300 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol) plus 0.3% NP40, 0.5 mM DTT, 10 μM MG132, 20 mM NEM and protease and phosphatase inhibitor cocktail (Thermo Scientific). Immunoprecipitations and immunoblotting were performed as previously described Torres et al.Citation23 Samples were resolved on a 4-20% gradient Tris gel (Bio-Rad) with MOPS running buffer, transferred to PVDF membrane and immunoblotted with indicated antibodies. Similarly, LAP-Tctex1d2 was expressed using .1 μg/ml doxycycline (Sigma-Aldrich) and tandem affinity purification and sample preparation for mass spectrometry were as described by Torres et al.Citation23 Mass spectrometry was conducted at the Harvard Mass Spectrometry and Proteomics Resource Laboratory by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Thermo LTQ-Orbitrap mass spectrometer as described by Vanderwerf et al.Citation33 or at the UCLA Pasarow Mass Spectrometry Laboratory on a Thermo LTQ-Orbitrap XL as described by Patananan et al.Citation34
Antibodies
Immunofluorescence, immunoblotting, and immunoprecipitations were carried out using antibodies against: GFP, Tctex1d2, Dync2h1, PCM1 (Abcam, cat# ab290, ab139804, ab122525, ab72443); Phospho-S10-H3 (Millipore, cat# 06570); Gapdh, S-tag (GeneTex, cat# GTX128060, GTX100118); α-tubulin (Serotec, cat# MCAP77); γ-tubulin, Ac-tubulin, Wdr60, CEP164, TTBK2 (Sigma, cat# T5326, T7451, HPA020607, HPA037605, HPA018113); Dync1h1, Dync1li2, Dynll1, Dynll2 and IFT88 (Proteintech, cat# 12345-1-AP, 18885-1-AP, 18130-1-AP, 16811-1-AP, 13967-1-AP); Wdr34 (Abgent, cat# AP12421C); Dync2li1 (Santa Cruz, cat# sc-376644). Centrin antibodies were a gift from J. Salisbury. Secondary antibodies conjugated to FITC, Cy3, and Cy5 were from Jackson Immuno Research and those conjugated to IRDye 680 and IRDye 800 were from LI-COR Biosciences.
Immunofluorescence microscopy
Immunofluorescence was carried out essentially as described by Torres et al.Citation32 Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100/PBS, and co-stained with 0.5 μg/ml Hoechst 33342 and indicated antibodies. Images were captured with a Leica DMI6000 microscope (Leica DFC360 FX Camera, 63x/1.40-0.60 NA oil objective, Leica AF6000 software) at room temperature using Hoechst 3342 and secondary FITC, Cy3, and Cy5 antibodies (Jackson Immuno). Images were deconvolved with Leica Application Suite 3D Deconvolution software and exported as TIFF files. Data represent the average ± SD of 3 independent experiments, with 100 cells counted for each.
In vitro binding assay
Full-length (amino acid residues 1-142), N-terminal (amino acid residues 1-40), and C-terminal (amino acid residues 41-142) TCTEX1D2 were fused to the C-terminus of the HA-tag to generate the pDEST-HA-TCTEX1D2-F, -N, and -C vectors. Similarly full-length WDR34 and WDR60 were fused to the C-terminus of the FLAG-tag to generate the pDEST-FLAG-WDR34/WDR60 vectors that were used for in vitro binding assays.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest.
985066_Supplementary_Materials.zip
Download Zip (1.7 MB)Funding
This material is based upon work supported by the National Science Foundation under Grant Number MCB1243645 to J.Z.T., any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. A P30 DK063491 grant to J.P.W also supported this work.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Waters AM, Beales PL. Ciliopathies: an expanding disease spectrum. Pediatr Nephrol 2011; 26:1039-56; PMID:21210154; http://dx.doi.org/10.1007/s00467-010-1731-7
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med 2011; 364:1533-43; PMID:21506742; http://dx.doi.org/10.1056/NEJMra1010172
- Huber C, Cormier-Daire V. Ciliary disorder of the skeleton. Am J Med Genet C Semin Med Genet 2012; 160C:165-74; PMID:22791528; http://dx.doi.org/10.1002/ajmg.c.31336
- McInerney-Leo AM, Schmidts M, Cortes CR, Leo PJ, Gener B, Courtney AD, Gardiner B, Harris JA, Lu Y, Marshall M, et al. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am J Hum Genet 2013; 93:515-23; PMID:23910462; http://dx.doi.org/10.1016/j.ajhg.2013.06.022
- Schmidts M, Vodopiutz J, Christou-Savina S, Cortes CR, McInerney-Leo AM, Emes RD, Arts HH, Tuysuz B, D'Silva J, Leo PJ, et al. Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am J Hum Genet 2013; 93:932-44; PMID:24183451; http://dx.doi.org/10.1016/j.ajhg.2013.10.003
- Patel-King RS, Gilberti RM, Hom EF, King SM. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol Biol Cell 2013; 24:2668-77; PMID:23864713; http://dx.doi.org/10.1091/mbc.E13-05-0266
- Rompolas P, Pedersen LB, Patel-King RS, King SM. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J Cell Sci 2007; 120:3653-65; PMID:17895364; http://dx.doi.org/10.1242/jcs.012773
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010; 328:593-9; PMID:20360068; http://dx.doi.org/10.1126/science.1181348
- Palmer KJ, MacCarthy-Morrogh L, Smyllie N, Stephens DJ. A role for Tctex-1 (DYNLT1) in controlling primary cilium length. Eur J Cell Biol 2011; 90:865-71; PMID:21700358; http://dx.doi.org/10.1016/j.ejcb.2011.05.003
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J Cell Biol 1998; 141:979-92; PMID:9585416; http://dx.doi.org/10.1083/jcb.141.4.979
- Korrodi-Gregorio L, Margarida Lopes A, Esteves SL, Afonso S, Lemos de Matos A, Lissovsky AA, da Cruz ESOA, da Cruz ESEF, Esteves PJ, Fardilha M. An intriguing shift occurs in the novel protein phosphatase 1 binding partner, TCTEX1D4: evidence of positive selection in a Pika model. PLoS One 2013; 8:e77236; PMID:24130861; http://dx.doi.org/10.1371/journal.pone.0077236
- Korrodi-Gregorio L, Vieira SI, Esteves SL, Silva JV, Freitas MJ, Brauns AK, Luers G, Abrantes J, Esteves PJ, da Cruz ESOA, et al. TCTEX1D4, a novel protein phosphatase 1 interactor: connecting the phosphatase to the microtubule network. Biol Open 2013; 2:453-65; PMID:23789093; http://dx.doi.org/10.1242/bio.20131065
- Torres JZ, Summers MK, Peterson D, Brauer MJ, Lee J, Senese S, Gholkar AA, Lo YC, Lei X, Jung K, et al. The STARD9/Kif16a Kinesin Associates with Mitotic Microtubules and Regulates Spindle Pole Assembly. Cell 2011; 147:1309-23; PMID:22153075; http://dx.doi.org/10.1016/j.cell.2011.11.020
- Pfister KK, Shah PR, Hummerich H, Russ A, Cotton J, Annuar AA, King SM, Fisher EM. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet 2006; 2:e1; PMID:16440056; http://dx.doi.org/10.1371/journal.pgen.0020001
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell 1999; 97:877-87; PMID:10399916; http://dx.doi.org/10.1016/S0092-8674(00)80800-4
- Mok YK, Lo KW, Zhang M. Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J Biol Chem 2001; 276:14067-74; PMID:11148215
- Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet 2006; 22:491-500; PMID:16860433; http://dx.doi.org/10.1016/j.tig.2006.07.006
- Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci U S A 2005; 102:3703-7; PMID:15738400; http://dx.doi.org/10.1073/pnas.0408358102
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development 2005; 132:1923-34; PMID:15790967; http://dx.doi.org/10.1242/dev.01775
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol 2005; 170:103-13; PMID:15998802; http://dx.doi.org/10.1083/jcb.200504008
- Smith JC, Northey JG, Garg J, Pearlman RE, Siu KW. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J Proteome Res 2005; 4:909-19; PMID:15952738; http://dx.doi.org/10.1021/pr050013h
- McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiol Genomics 2008; 32:198-206; PMID:17971504; http://dx.doi.org/10.1152/physiolgenomics.00128.2007
- Torres JZ, Miller JJ, Jackson PK. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 2009; 9:2888-91; PMID:19405035; http://dx.doi.org/10.1002/pmic.200800873
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498-504; PMID:14597658; http://dx.doi.org/10.1101/gr.1239303
- Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N. Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet 2003; 72:650-8; PMID:12567324; http://dx.doi.org/10.1086/368204
- Brancati F, Iannicelli M, Travaglini L, Mazzotta A, Bertini E, Boltshauser E, D'Arrigo S, Emma F, Fazzi E, Gallizzi R, et al. MKS3/TMEM67 mutations are a major cause of COACH Syndrome, a Joubert Syndrome related disorder with liver involvement. Hum Mutat 2009; 30:E432-42; PMID:19058225; http://dx.doi.org/10.1002/humu.20924
- Mohan S, Timbers TA, Kennedy J, Blacque OE, Leroux MR. Striated rootlet and nonfilamentous forms of rootletin maintain ciliary function. Curr Biol 2013; 23:2016-22; PMID:24094853; http://dx.doi.org/10.1016/j.cub.2013.08.033
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 2007; 179:321-30; PMID:17954613; http://dx.doi.org/10.1083/jcb.200707181
- Goetz SC, Liem KF, Jr., Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 2012; 151:847-58; PMID:23141541; http://dx.doi.org/10.1016/j.cell.2012.10.010
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 2000; 151:709-18; PMID:11062270; http://dx.doi.org/10.1083/jcb.151.3.709
- Merrill AE, Merriman B, Farrington-Rock C, Camacho N, Sebald ET, Funari VA, Schibler MJ, Firestein MH, Cohn ZA, Priore MA, et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am J Hum Genet 2009; 84:542-9; PMID:19361615; http://dx.doi.org/10.1016/j.ajhg.2009.03.015
- Torres JZ, Ban KH, Jackson PK. A specific form of phospho protein phosphatase 2 regulates anaphase-promoting complex/cyclosome association with spindle poles. Mol Biol Cell 2010; 21:897-904; PMID:20089842; http://dx.doi.org/10.1091/mbc.E09-07-0598
- Vanderwerf SM, Svahn J, Olson S, Rathbun RK, Harrington C, Yates J, Keeble W, Anderson DC, Anur P, Pereira NF, et al. TLR8-dependent TNF-(alpha) overexpression in Fanconi anemia group C cells. Blood 2009; 114:5290-8; PMID:19850743; http://dx.doi.org/10.1182/blood-2009-05-222414
- Patananan AN, Capri J, Whitelegge JP, Clarke SG. Non-repair pathways for minimizing protein isoaspartyl damage in the yeast saccharomyces cerevisiae. J Biol Chem 2014; 289:16936-53; PMID:24764295; http://dx.doi.org/10.1074/jbc.M114.564385
