Abstract
Mitochondrial DNA (mtDNA) is a multi-copy genome encoding for proteins essential for aerobic energy metabolism. Mutations in mtDNA can lead to a variety of human diseases, from mild metabolic syndromes to severe fatal encephalomyopathies. Most mtDNA mutations co-exist with wild type genomes in a state known as heteroplasmy. The segregation of these pathogenic mutants is tissue and mutation specific, and a key determinant in the onset and severity of human mitochondrial disorders. We used a forward genetic approach in mice to identify and demonstrate that Gimap3 (GTPase of immunity associated protein) is a key regulator of mtDNA segregation in leukocytes. The Gimap gene cluster is found only in vertebrates and appear to be a class of nucleotide-dependent dimerization GTPases. Gimap3 is a membrane-anchored GTPase with a critical role in T cell development. Here, we summarize our genetic findings and postulate how Gimap3 might regulate mtDNA genetics.
Mitochondrial DNA (mtDNA) is a maternally inherited circular genome of approximately 16 kb with a copy number that ranges from 102 and 105 per cell.Citation1,Citation2 Typically, an individual or cell contains only one variant or haplotype of the mitochondrial genome, a state known as homoplasmy. Somatic and germ line mutations can cause two or more variants to co-exist (heteroplasmy) and can impair mitochondrial oxidative phosphorylation when they exceed a threshold. Segregation of heteroplasmic mtDNA variants is usually a random process in both the germline and in somatic cells, dependent on mtDNA turnover and copy number.Citation3 However, some pathogenic mtDNA mutations can have skewed segregation patterns affecting the onset and severity of disease. These segregation patterns are mutation and tissue-specific but independent of the biochemical defect. Two of the more common mitochondrial tRNA mutations in tRNALeu (OMIM:590050) and tRNALys (OMIM:590060) illustrate the point. Both mutations impair translation of mitochondrially-encoded proteins, yet genetically are treated differently in leukocytesCitation4,Citation5 by unknown mechanisms.
To identify the molecular basis for tissue-specific mtDNA segregation, we used a heteroplasmic mouse model combined with a classical forward genetics approach to map and clone genes regulating this phenotype. This mouse model carries two neutral mtDNA variants derived from two old inbred mouse strains BALB and NZB, and display age-dependent and tissue-specific selection for mtDNA haplotypes.Citation6 In hematopoietic tissues, there is selection for BALB mtDNA, a phenotype maintained on a number of different mouse nuclear backgrounds.Citation7 We set up a genetic cross with the more distantly related subspecies Mus domesticus castaneus, to map an 11 Mb locus on chromosome 6 as a key regulator for mtDNA segregation in mouse hematopoietic tissues.Citation7,Citation8 Within this locus we identified a mutation in the splice acceptor site of the castaneus Gimap3 (GTPase of immunity associated protein) variant that alters mRNA splicing, adding an extra 58 amino acids to the mature protein (). We made transgenic mice overexpressing this CAST Gimap3 cDNA and could decrease the rate of mtDNA segregation in the spleen.Citation8 This work identified the first nuclear-encoded gene to regulate mtDNA genetics in a tissue-specific manner.
Gimap3 is a 34 kDa protein containing a AIG domain (Pfam: PF04548), from the Arabidopsis AIG1 originally identified in resistance to bacterial invasion and considered the prototype for the Gimap family in vertebrates.Citation9 Generally, this domain consists of a G1–G5 GTPase domain and a hydrophobic conserved box located between G3 and G4. In addition, Gimap3 contains a putative coiled-coil domain and a C-terminal transmembrane domain for membrane anchoring. The CAST Gimap3 variant has an extra 58 amino acids at the N-terminus of the protein (41 kDa), while the core of the protein is identical including its transmembrane domains ().Citation8 Gimap3 is only expressed in leukocytes, consistent with our mtDNA genetic phenotype, although absent in some mouse leukemia lines.Citation10
The most comprehensive knowledge for members of the Gimap family comes from studies on Gimap5, a Gimap3 paralogue 84% identical at the amino acid level, differing only at the immediate N-terminus and critically at the C-terminus. Gimap5 was first identified as a null mutant in the BioBreeding rat model for type I diabetes, which develop a severe T cell lymphopenia and a prerequisite to the autoimmunity.Citation11 In mice, loss of Gimap5 has far more deleterious consequences producing a severe defect in hematopoiesis affecting T and B lymphocytes, NK cells, granulocytes and erythropoiesis.Citation12,Citation13 In mice, T lymphocyte development and survival requires both Gimap3 and Gimap5, which are upregulated in the transition from immature double positive thymocytes to single positive (CD4+ and CD8+) T cells in the thymus.Citation10 This expression pattern is similar to that observed for Bcl2 in T cells,Citation14 and not surprising, both Gimaps interact with Bcl2 family members (anti- and pro-apoptotic).Citation10 The molecular basis for Gimap3 and Gimap5 interactions with Bcl2 family members and their specific requirement during T cell maturation and survival is an open question that needs addressing.
The high identity of Gimap3 and Gimap5 with a similar expression profile in leukocytes, would suggest overlapping functions. However, it is unlikely that these functions are at the same intracellular location, because these proteins differ significantly at the C-terminal transmembrane domain. Membrane insertion and localization are determined by the transmembrane domain and flanking sequence at the C-terminus. The original papers describing Gimap3 and Gimap5, assigned these proteins to a variety of intracellular membranes including mitochondria, endoplasmic reticulum and Golgi among others, using tagged constructs and massive overexpression perhaps leading to some mislocalization.Citation15,Citation16 To illustrate the point, a recent paper on Gimap5 localization is more convincing,Citation16 using a series of species-specific monoclonal antibodies, to demonstrate insertion into lysosomes and multivesicular bodies, which is at odds from earlier findings.Citation15 In light of this finding, the localization for Gimap3 needs to be revisited.
A big step in elucidating the function for Gimap family members was made recently with the crystal structure for GIMAP2, which undergoes GTP-dependent oligomerization to increase intracellular lipid droplet formation.Citation17 Gimaps appear to belong to a distinct cluster of TRAFAC (translation factor associated) GTPases containing Tocs (translocon at the outer envelope membrane of chloroplasts), septins and dynamin.Citation17 Based upon the structural similarities of Gimaps, it now appears this protein family can form nucleotide-dependent oligomers either as scaffolds for interacting partners on membranes or for membrane remodeling. Oligomerization of Gimap3 and Gimap5 could affect their reported interaction with Bcl2 family members and their role in apoptosis at different intracellular membranes. Likewise, the CAST Gimap3 variant could be impaired in its ability to oligomerize at the G interface, and thus unable to form scaffolds.
How might the intracellular segregation of mtDNA be coordinated? It has long been thought that mechanisms selecting for different mtDNA haplotypes or mutants must be a consequence of respiratory chain function, mtDNA replication rates or copy number regulation. This is clearly not the case for all human pathogenic mtDNA mutations, as segregation patterns are mutation and tissue-specific, even though they all lead to a defect in aerobic energy metabolism. We have empirically tested these factors in our heteroplasmic mice and found none can modulate the selective pressure on mtDNA haplotypes.Citation8,Citation18,Citation19 Nonetheless, mtDNA segregation can only occur at two levels, on the mtDNA sequence itself or the encoded polypeptides, so how might it happen?
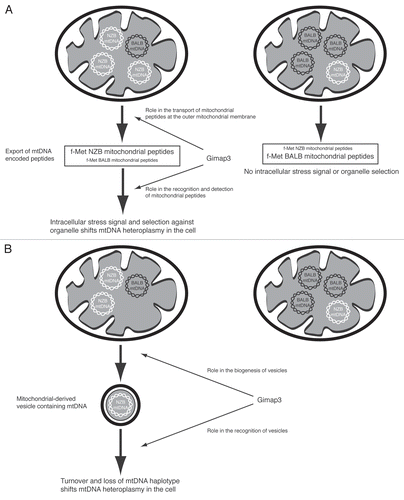
Mitochondria are derived from an ancient bacterium and still maintain some functions with more similarity to their ancestors, such as genome organization and protein translation. Since mitochondrial contents are generally hidden behind one to two lipid bilayers, sequestered away from the cytosolic environment, it is not surprising that release of these contents can have devastating effects on cellular homeostasis, cytochrome c being the most glaring example. One way to identify mtDNA haplotypes is through the polypeptides they encode, which could act as intracellular signals. Mitochondrial peptide export is a conserved cellular process, first described for the maternally-inherited antigen,Citation20 and then subsequently found in S. cerevisiaeCitation21 and in C. elegansCitation22 where it is involved in a stress response. In mice, mtDNA-encoded polypeptides with a formyl-methionine were selectively exported from the organelle and form the basis for priming the adaptive immune system against Listeria infection. Interestingly, the NZB mtDNA haplotype was critical in these experiments,Citation20 and in our heteroplasmic mouse model, leukocytes select against this same mitochondrial genome in favor of the BALB mtDNA haplotype. However, when we genetically disrupted adaptive immunity in mice, there was no effect on mtDNA segregation.Citation19 Although, a recent report demonstrated that in addition to the adaptive immune system, the innate immune system, via NK cells, can also recognize and kill cells harboring distinct mtDNA haplotypes, particularly NZB mtDNA.Citation23 The mechanism for distinguishing mtDNA haplotypes is unknown and what happens with a heteroplasmic situation. One possibility could be when leukocytes are heteroplasmic, a biased export of polypeptides encoded by NZB mtDNA elicits an intracellular stress response to reduce the proportion of this haplotype in the cell. These peptides could be a signal for identifying and selecting particular mitochondrial genomes ().
Mitochondrial organelle shape and movement are very dynamic and cell-specific, coordinated by a series of GTPases on the outer and inner mitochondrial membrane.Citation24 In leukocytes, mitochondrial shape is not reticulated, as commonly seen in cultured adherent cell lines, instead it is small and globular, and can be reorganized in response to chemotaxis and cell activation.Citation25,Citation26 These outer mitochondrial membrane GTPases also have key roles stabilizing the mitochondrial-endoplasmic reticulum interface required for mitochondrial calcium homeostasis,Citation27 and in Bax-dependent apoptosis.Citation28 Recently, mitochondrial-derived vesicles have been described that bud off of the mitochondrial reticulum. These vesicles form by a dedicated protein machinery leading to cargo selectivity and fusion with peroxisomes.Citation29 This mechanism is independent of Drp1 (Dynamin-related protein 1), the master regulator of mitochondrial fission. To date, mtDNA has not been detected in these vesicles, however, eubacteria possess an analogous cargo system, where vesicles contain plasmid DNA.Citation30 Whether mitochondrial-derived vesicles could form and fuse with other intracellular organelle compartments, such as endosomes or lysosomes, as a means to control mtDNA turnover remains to be determined. Nonetheless, it is tempting to speculate on Gimap3 involvement in some retromer vesicle pathway, either at the site of formation or with fusion to other organelles, where the CAST variant would disrupt the process (). Such a system, would still require a mechanism, likely tissue-specific, to distinguish between mtDNA haplotypes and packaging mtDNA into vesicles, so many questions remain as to the feasibility of this hypothesis.
The discovery that Gimap3 can regulate mtDNA genetics and likely functions as a membrane scaffold or in membrane remodeling opens up interesting new biology to regulate the genetics of mtDNA. Functionally, Gimap3 might intersect with cargo selective vesicles budding from mitochondria. Future work on Gimap3 and Gimap5 function will lead the way to elucidating their role in regulating mitochondrial genetics and involvement in mitochondrial retromer pathways.
Figures and Tables
Figure 1 Gimap3 protein variation in mice. (A) Schematic of BALB/c and CAST/Ei Gimap3 variants with an AIG domain, coiled-coil (CC) and transmembrane domain (TM). The AIG domain consists of a G1–G5 GTPase domain and a hydrophobic conserved box located between G3 and G4. BALB and CAST Gimap3 variants are identical in sequence except at the N-terminus due to differential splicing resulting in an additional 58 amino acids in the CAST form. All common laboratory mouse strains (Mus mus domesticus) have the BALB variant of Gimap3. (B) Amino acid sequence alignment at the N-terminus of BALB and CAST Gimap3. Black boxes indicate regions of identity and the beginning of the AIG domain is indicated.

Figure 2 Possible models for selecting mitochondrial DNA (mtDNA) haplotypes in leukocytes and the potential role of Gimap3. (A) MtDNA selection is due to intracellular organelle selection resulting from the export of NZB-encoded mitochondrial peptides. Mitochondrially-encoded fMet peptides are known to be exported from mitochondria and form the basis of the maternally-transmitted antigen. Gimap3 could function in the export or recognition of mitochondrially-encoded peptides. (B) MtDNA selection is due to mitochondrially-derived vesicles. Vesicles are known to bud off from mitochondria. Gimap3 could function in the biogenesis or recognition of vesicles.
Acknowledgements
This work was supported by grants B.J.B. from the Academy of Finland, University of Helsinki, Biocentrum Helsinki and UMDF-USA. R.J. is supported by the Helsinki Biomedical Graduate School. B.J.B. is an Academy of Finland Research Fellow.
Extra View to: Jokinen R, Marttinen P, Sandell HK, Manninen T, Teerenhovi H, Wai T, et al. Gimap3 regulates tissue-specific mitochondrial DNA segregation. PLoS Genet 2010; 6:e1001161; PMID: 20976251; http://dx.doi.org/10.1371/journal.pgen.1001161
References
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet 2005; 6:389 - 402
- Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet 2008; 40:1484 - 1488
- Chinnery PF, Samuels DC. Relaxed replication of mtDNA: A model with implications for the expression of disease. Am J Hum Genet 1999; 64:1158 - 1165
- Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 A→G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet 2001; 68:238 - 240
- Rajasimha HK, Chinnery PF, Samuels DC. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A→G mutation in blood. Am J Hum Genet 2008; 82:333 - 343
- Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet 1997; 16:93 - 95
- Battersby BJ, Loredo-Osti JC, Shoubridge EA. Nuclear genetic control of mitochondrial DNA segregation. Nat Genet 2003; 33:183 - 186
- Jokinen R, Marttinen P, Sandell HK, Manninen T, Teerenhovi H, Wai T, et al. Gimap3 regulates tissue-specific mitochondrial DNA segregation. PLoS Genet 2010; 6:1001161
- Krucken J, Schroetel RM, Muller IU, Saidani N, Marinovski P, Benten WP, et al. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene 2004; 341:291 - 304
- Nitta T, Nasreen M, Seike T, Goji A, Ohigashi I, Miyazaki T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol 2006; 4:103
- Hornum L, Romer J, Markholst H. The diabetesprone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes 2002; 51:1972 - 1979
- Schulteis RD, Chu H, Dai X, Chen Y, Edwards B, Haribhai D, et al. Impaired survival of peripheral T cells, disrupted NK/NKT cell development and liver failure in mice lacking Gimap5. Blood 2008; 112:4905 - 4914
- Barnes MJ, Aksoylar H, Krebs P, Bourdeau T, Arnold CN, Xia Y, et al. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. J Immunol 2010; 184:3743 - 3754
- Veis DJ, Sentman CL, Bach EA, Korsmeyer SJ. Expression of the Bcl-2 protein in murine and human thymocytes and in peripheral T lymphocytes. J Immunol 1993; 151:2546 - 2554
- Sandal T, Aumo L, Hedin L, Gjertsen BT, Doskeland SO. Irod/Ian5: an inhibitor of gamma-radiation- and okadaic acid-induced apoptosis. Mol Biol Cell 2003; 14:3292 - 3304
- Wong VWYSA, Hutchings A, Pascall JC, Carter C, Bright N, Walker SA, et al. The autoimmunity-related GIMAP5 GTPase is a lysosome-associated protein. Self/Nonself 2010; 1:259 - 268
- Schwefel D, Frohlich C, Eichhorst J, Wiesner B, Behlke J, Aravind L, et al. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2). Proc Natl Acad Sci USA 2010; 107:20299 - 20304
- Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum Mol Genet 2001; 10:2469 - 2479
- Battersby BJ, Redpath ME, Shoubridge EA. Mitochondrial DNA segregation in hematopoietic lineages does not depend on MHC presentation of mitochondrially encoded peptides. Hum Mol Genet 2005; 14:2587 - 2594
- Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell 1990; 60:971 - 980
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science 2001; 291:2135 - 2138
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 2010; 37:529 - 540
- Ishikawa K, Toyama-Sorimachi N, Nakada K, Morimoto M, Imanishi H, Yoshizaki M, et al. The innate immune system in host mice targets cells with allogenic mitochondrial DNA. J Exp Med 2010; 207:2297 - 2305
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem 2007; 76:751 - 780
- Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med 2006; 203:2879 - 2886
- Quintana A, Schwindling C, Wenning AS, Becherer U, Rettig J, Schwarz EC, et al. T cell activation requires mitochondrial translocation to the immunological synapse. Proc Natl Acad Sci USA 2007; 104:14418 - 14423
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008; 456:605 - 610
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol 2007; 177:439 - 450
- Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol 2008; 18:102 - 108
- Renelli M, Matias V, Lo RY, Beveridge TJ. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 2004; 150:2161 - 2169