Abstract
Ras proteins are best known to function on the plasma membrane to mediate growth factor signaling. Controlling the length of time that Ras proteins stay on the plasma membrane is an effective way to properly modulate the intensity and duration of growth factor signaling. It has been shown previously that H- and N-Ras proteins in the GTP-bound state can be ubiquitylated via a K-63 linkage, which leads to endosome internalization and results in a negative-feedback loop for efficient signal attenuation. In a more recent study, two new Ras effectors have been isolated, CHMP6 and VPS4A, which are components of the ESCRT-III complex, best known for mediating protein sorting in the endosomes. Surprisingly, these molecules are required for efficient Ras-induced transformation. They apparently do so by controlling recycling of components of the Ras pathway back to the plasma membrane, thus creating a positive-feedback loop to enhance growth factor signaling. These results suggest the fates of endosomal Ras proteins are complex and dynamic — they can be either stored and/or destroyed or recycled. Further work is needed to decipher how the fate of these endosomal Ras proteins is determined.
Humans have three RAS genes, H-, N- and K-RAS and because the latter can undergo alternative splicing to yield two isoforms (K-Ras-4A and K-Ras-4B), a total of four Ras proteins can be found in many cells. Ras proteins cycle between the GDP and GTP-bound states to act as binary switches to control a wide range of signal transduction pathways. Mutations that render Ras proteins constitutively GTP-bound are among the most frequent genetic alterations in human cancers—approximately 30% of them carrying an oncogenic RAS mutationCitation1 (and data from the COSMIC database, www.sanger.ac.uk/perl/genetics/CGP/cosmic). The best known Ras function relevant to tumor formation is the control of growth factor signaling, which occurs at the plasma membrane. While this landmark finding provides a satisfactory explanation of how oncogenic Ras can induce tumorigenesis, many important questions remain largely unanswered. For example, how do Ras proteins get to the plasma membrane, and once there, what eventually happens to them? Most Ras proteins also accumulate in the cytoplasm (see below), are these proteins active in signaling?
On and Off the Plasma Membrane
While nascent Ras polypeptides are soluble and cytoplasmic, all Ras proteins contain a C-terminal CAAX motif whose cysteine can be farnesylated. The “AAX” is then cleaved off by a protease, and the farnesylated cysteine is later methylated. Ras proteins that are modified in this manner gain general affinity to the cell membrane, but for most Ras proteins, except K-Ras-4B, the more abundant and ubiquitously expressed K-Ras isoform, they cannot efficiently associate with the plasma membrane, unless they are further palmitoylated at cysteine(s) just upstream of the CAAX motif. The presumptive Ras acyl palmitoyltransferase zDHHC9 is found in the Golgi,Citation2 supporting the concept that most Ras proteins use the Golgi as a launching pad to reach the plasma membrane via the trans-Golgi system.Citation3,Citation4 Instead of palmitoylation, K-Ras-4B associates with the plasma membrane via electrostatic interactions with a lysine rich region in the C-terminus. However, how K-Ras-4B localizes to the plasma membrane remains largely unknown and there is no evidence that it does so by the trans-Golgi system.Citation5
What happens to Ras proteins once they reach the plasma membrane is complex and not fully resolved. The palmitoylation is reversible and the palmitoyl moiety can be removed by acyl protein thioesterase (APT1).Citation6 De-palmitoylated Ras proteins, by mechanisms that are not fully understood, readily accumulate in the Golgi, awaiting another round of palmitoylation.Citation7,Citation8 This palmitoylation/depalmitoylation cycle can evidently act as an ON-OFF switch to control signaling at the plasma membrane.
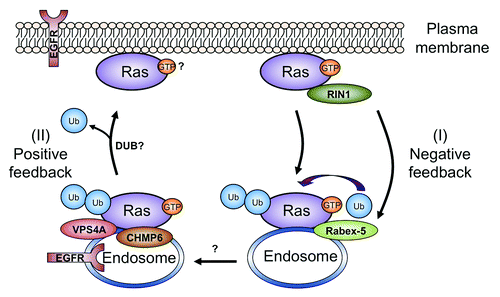
In addition to Golgi localization, H- and N-Ras have also been shown to accumulate in endosomesCitation9 (). One key role of endosomes in signaling is illustrated by a process known as receptor-mediated endocytosis, in which activated receptors are internalized (via endocytosis) to reside in early endosomes.Citation10 For efficient signaling attenuation, these receptors are transferred to late endosomes and ultimately delivered to lysosomes for degradation. Intriguingly, RIN1, a Ras effector,Citation11 acts as a guanine nucleotide exchange factor for Rab5,Citation11,Citation12 which is a key component for endocytosis. This suggests that Ras may attenuate its own signaling by promoting internalization of either growth factor receptors (e.g., EGFR) or Ras itself. Evidence for the latter was provided by the discovery that H- and N-Ras proteins are both mono- and di-ubiquitylated via a K63-linkage, a mode of ubiquitylation that is often required for endocytosis as well as protein sorting at the endosome.Citation13 When Ras proteins are deficient in ubiquitylation by either mutating the lysine residues in RasCitation13 or by silencing the Ras E3 ligase Rabex-5/RabGEF1,Citation14,Citation15 the Ras signaling outputs are enhanced. Conversely, when Rabex-5 is overexpressed, Ras signaling is inhibited.Citation14,Citation15 These results suggest that a key outcome of Ras internalization is to attenuate growth factor signaling. Despite this, several lines of evidence suggests that internalized EGFR and/or Ras may be active. For example, EGFR remains EGF-bound and phosphorylated in early endosomes,Citation16 and Ras can also interact with PI3K and Cdc42 in endosomes.Citation17,Citation18 Collectively, it seems plausible that the fate of endosomal Ras is quite complex and that not all the internalized Ras proteins are destined for storage or destruction.
Figure 1. A model of the shuttling of H-Ras or N-Ras between the plasma membrane and endosomes. Activated Ras proteins stimulate the RIN1 effector, which then activates and recruits Rabex-5 to ubiquitylate Ras to facilitate retention in endosomes.Citation14 One consequence of endosomal internalization is suppression of the signaling output from the Ras pathway (I).Citation13–Citation15 A fraction of the GTP-bound and ubiquitylated Ras can also stimulate ESCRT-III components CHMP6 and VPS4A to enable sorting and recycling of components from the Ras pathway, such as EGFR (and possibly Ras itself) back to the plasma membrane. This pathway apparently enhances Ras signaling (II).Citation28 Ubiquitylation of Ras in Pathway I forms a negative feedback loop to more efficiently attenuate Ras signaling, while Pathway II acts in a positive feedback loop for sustained and prolonged Ras signaling. Proper balancing of these two pathways can serve to fine tune Ras signaling outputs. Question marks denote many important questions that hopefully will be addressed in the future (see text).
Ras Signaling From the Cytoplasm
At steady-state, H- and N-Ras proteins can be readily detected in the cytoplasmic endomembranes (e.g., endosomes, ER and Golgi, etc). Are these Ras proteins inactive as “immature” covalent modification intermediates, internalized and destined for degradation, or do they activate cytoplasmic effectors necessary for cell transformation?
One of the earliest reports on Ras signaling from the cytoplasm came from Hancock et al., who showed that when oncogenic H-Ras is restricted in the cytoplasm by mutations that abolish palmitoylation, the resulting protein can still transform cells.Citation19 Later, Chiu et al. more systematically investigated this by specifically targeting oncogenic H-Ras to the ER and Golgi and found that these proteins can efficiently transform NIH3T3 cells.Citation20 A Golgi specific Ras pathway in response to viral infection has also been found in T-cells.Citation21 In addition to ER and Golgi, Ras proteins have been shown to function in mitochondria to control apoptosis and glucose metabolism.Citation22,Citation23 Despite the evidence for compartmentalized Ras signaling, specific Ras effectors for a given cell compartment are largely unknown in mammalian cells. However, we have shown that in the fission yeast Schzosaccharomyces pombe, a single Ras protein, Ras1, controls two spatially segregated pathways.Citation24–Citation26 On the plasma membrane, Ras1 controls Byr2, a MEK-like protein kinase, to mediate mating pheromone signaling. On the endomembrane, Ras stimulates Scd1, a nucleotide exchange factor for Cdc42, to mediate cell morphogenesis. Guided by the Ras-Cdc42 interaction in S. pombe, Cheng et al. have recently determined in mammalian cells that H- and N-Ras also activate Cdc42 in the endomembrane (e.g., endosomes), an interaction that is critical for Ras-induced transformation.Citation18,Citation27
Since most compartmentalized Ras effectors are unknown, in a soon to be published paper, Zheng et al. have conducted a screen to identify new Ras effectors and to categorize them based on where in the cell they interact with Ras.Citation28 To achieve this goal, they used a microscopy-based technology called Biomolecular Fluorescence Complementation (BiFC), in which an N- and C-terminal fragment of YFP (Yn and Yc) are each fused to a protein, and a fluorescently competent YFP is reconstituted when the fused proteins form a complex.Citation29 Using oncogenic H-Ras as bait, FACS was first employed to allow high throughput screening of a human cDNA library for Ras binding proteins, followed by regular fluorescent microscopy to analyze where the binding takes place in the cell. Promising candidate clones were also screened functionally by the ability to alter Ras-induced activities including transformation. Interestingly, of the 26 final candidate effectors, more than 1/3 are known to regulate protein transport, two of which, CHMP6/VPS20 and VPS4A, are well-known ESCRT-III (Endosomal Sorting Complex Required for Transport-III) componentsCitation30 and were chosen for detail analysis.
Escorting Ras Back to the Plasma Membrane
A key role of the ESCRT-III components is to promote scission of the intraluminal vesicles as endosomal cargos are sorted into different compartments (e.g., the multivesicular bodies/late endosomes). In this study, CHMP6 and VPS4A are classic Ras effectors in that they bind H-Ras directly and the binding is GTP-dependent, while H-Ras acts as a conventional endosome cargo in that it needs to be ubiquitylated for the binding. By microscopy and marker analysis, the binding was mapped to early and late endosomes, but not recycling endosomes or Golgi. Despite the fact that earlier studies suggest that internalized Ras proteins may be headed for a more dormant state, surprisingly, when expression of CHMP6 or VPS4A is repressed, Ras-induced transformation is concurrently attenuated. To determine the reason for this, biochemical fractionation experiments were performed and showed that in CHMP6 or VPS4A repressed cells, the pool of Ras proteins on the plasma membrane is reduced. Furthermore, by photobleaching experiments, silencing CHMP6 or VPS4A greatly reduced Ras movement from the cytoplasm to the plasma membrane. Taken together, these data suggest that CHMP6 and VPS4A control recycling of Ras and/or components of the Ras pathway back to the plasma membrane. EGFR recycling is well known to be controlled by ESCRT-III.Citation31 Indeed, Zheng et al. present evidence that Ras can act through CHMP6 and VPS4A to control EGFR cycling. Thus while one of the key roles of Ras internalization is no doubt to attenuate growth factor signaling, the study by Zheng et al. offers an alternative (). That is, Ras proteins can also stimulate CHMP6 and VPS4A to induce recycling of Ras proteins themselves and/or other key components of the Ras pathway, such as EGFR, to create a positive feedback loop for sustained growth factor signaling.
Concluding Remarks
It is becoming clear that a unidirectional flux of nascent Ras proteins streaming from the cytoplasm to the plasma membrane as they become covalently modified is unlikely to explain what is really occurring at the plasma membrane. It seems likely that there are at least three pools of Ras proteins at the plasma membrane: newly synthesized and covalently modified arriving from the Golgi, and recycled via the palmitoylation/depalmitoylation cycle, and recycled via the ubiquitylation cycle. The contribution of each of these populations to the total plasma membrane pool remains to be determined.
We note that there are still many important unanswered questions regarding Ras in the endosomes. It is generally accepted that endosomal cargos need to shed their ubiquitin code in order to exit the endosomes, and this process is performed by the so called deubiquitinating enzymes, or DUBs (). Identification of these Ras-specific DUBs may shed light on the role of deubiquitylation in the control of Ras recycling back to the plasma membrane. One of the best known fates of proteins in the late endosomes is to be sent to the lysosome for degradation, and there is evidence that K-Ras can be degraded in the lysosome.Citation32 How do Ras proteins in late endosomes avoid being degraded? Does this process require interaction with CHMP6, VPS4A and some yet to be identified sorting factors to channel Ras proteins into the recycling route or does de-ubiquitylation (by DUBs) effectively block delivery to the lysosome? Finally, it is unclear whether the recycled Ras proteins are still GTP bound, which, conceivably, can readily stimulate effectors on the plasma membrane.
As noted earlier that K-Ras-4B is a “black sheep” in terms of how it associates with the plasma membrane because it is not palmitoylated. The different modes by which different Ras proteins associate with the plasma membrane appear to influence how Ras proteins localize in the cell. While cytoplasmic N- and H-Ras can be readily found in the cytoplasm, K-Ras-4B is usually exclusively found on the plasma membrane. Consistent with this, many cytoplasmic Ras effectors preferentially interact with H- and N-Ras but not with K-Ras-4B. In Cheng et al., Cdc42 has been shown to interact with only H- and N-Ras, but not with K-Ras-4B.Citation18 In the current study by Zheng et al., CHMP6 and VPS4A also only bind H- and N-Ras, but not K-Ras-4B, in the cell lines that were examined. Furthermore, when CHMP6 or VPS4A was repressed, EGFR recycling was impeded in cancer cell lines carrying oncogenic N-RAS but not oncogenic K-RAS.Citation28 Collectively, these data suggest that the observed interaction with CHMP6 and VPS4A is a unique feature for N- and H-Ras. This may be partly explained by the fact that while H- and N-Ras are ubiquitylated via a K-63 linkage,Citation13 the ubiquitylation of K-Ras-4B appears primarily through a K-48 linage.Citation33 In human cancer, K-RAS oncogenic mutations are found in a wide range of tissues, such as pancreas (57%), large (33%) and small (20%) intestine, biliary tract (31%), lung (17%), etc.Citation34 By contrast, oncogenic mutations in H- and N-RAS are more tissue specific. For example, oncogenic N-RAS mutations are common in tumors of the skin (18%) and hematopoietic tissues (10%); while oncogenic H-RAS mutations are frequently found in tumors in the head and neck (e.g., salivary gland (15%), upper aerodigestive tract (9%), etc.).Citation34 The molecular mechanisms for the apparent Ras isoform-specific roles in cancer remain largely unclear. However, one possibility, as uncovered by the current study of Zheng et al. and by other studies, is that different Ras proteins, diversified further by covalent modifications, can interact with a different set of effectors during tumorigenesis.
Acknowledgments
ZZ is supported by a postdoctoral fellowship from the Susan G. Komen for the Cure Foundation (PDF0707860). DB-S is supported by grants from NIH (CA055360 and GM078266). ECC is supported by grants from NIH (CA90464, CA107187, GM81627 and P50-CA58183).
References
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res 1989; 49:4682 - 9; PMID: 2547513
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, et al. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem 2005; 280:31141 - 8; http://dx.doi.org/10.1074/jbc.M504113200; PMID: 16000296
- Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol 2000; 20:2475 - 87; http://dx.doi.org/10.1128/MCB.20.7.2475-2487.2000; PMID: 10713171
- Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol 2010; 191:23 - 9; http://dx.doi.org/10.1083/jcb.200911143; PMID: 20876282
- Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 2012; 13:39 - 51; http://dx.doi.org/10.1038/nrm3255; PMID: 22189424
- Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J Biol Chem 1998; 273:15830 - 7; http://dx.doi.org/10.1074/jbc.273.25.15830; PMID: 9624183
- Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol 2005; 170:261 - 72; http://dx.doi.org/10.1083/jcb.200502063; PMID: 16027222
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 2005; 307:1746 - 52; http://dx.doi.org/10.1126/science.1105654; PMID: 15705808
- Gomez GA, Daniotti JL. H-Ras dynamically interacts with recycling endosomes in CHO-K1 cells: involvement of Rab5 and Rab11 in the trafficking of H-Ras to this pericentriolar endocytic compartment. J Biol Chem 2005; 280:34997 - 5010; http://dx.doi.org/10.1074/jbc.M506256200; PMID: 16079139
- Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol 2011; 23:393 - 403; http://dx.doi.org/10.1016/j.ceb.2011.03.008; PMID: 21474295
- Han L, Wong D, Dhaka A, Afar D, White M, Xie W, et al. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci U S A 1997; 94:4954 - 9; http://dx.doi.org/10.1073/pnas.94.10.4954; PMID: 9144171
- Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell 2001; 1:73 - 82; http://dx.doi.org/10.1016/S1534-5807(01)00008-9; PMID: 11703925
- Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell 2006; 21:679 - 87; http://dx.doi.org/10.1016/j.molcel.2006.02.011; PMID: 16507365
- Xu L, Lubkov V, Taylor LJ, Bar-Sagi D. Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr Biol 2010; 20:1372 - 7; http://dx.doi.org/10.1016/j.cub.2010.06.051; PMID: 20655225
- Yan H, Jahanshahi M, Horvath EA, Liu HY, Pfleger CM. Rabex-5 ubiquitin ligase activity restricts Ras signaling to establish pathway homeostasis in Drosophila. Curr Biol 2010; 20:1378 - 82; http://dx.doi.org/10.1016/j.cub.2010.06.058; PMID: 20655224
- Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell 2001; 12:1897 - 910; PMID: 11408594
- Tsutsumi K, Fujioka Y, Tsuda M, Kawaguchi H, Ohba Y. Visualization of Ras-PI3K interaction in the endosome using BiFC. Cell Signal 2009; 21:1672 - 9; http://dx.doi.org/10.1016/j.cellsig.2009.07.004; PMID: 19616621
- Cheng CM, Li H, Gasman S, Huang J, Schiff R, Chang EC. Compartmentalized Ras proteins transform NIH 3T3 cells with different efficiencies. Mol Cell Biol 2011; 31:983 - 97; http://dx.doi.org/10.1128/MCB.00137-10; PMID: 21189290
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 1989; 57:1167 - 77; http://dx.doi.org/10.1016/0092-8674(89)90054-8; PMID: 2661017
- Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol 2002; 4:343 - 50; PMID: 11988737
- Perez de Castro I, Bivona TG, Philips MR, Pellicer A. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol Cell Biol 2004; 24:3485 - 96; http://dx.doi.org/10.1128/MCB.24.8.3485-3496.2004; PMID: 15060167
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell 2006; 21:481 - 93; http://dx.doi.org/10.1016/j.molcel.2006.01.012; PMID: 16483930
- Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y, et al. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res 2012; 22:399 - 412; http://dx.doi.org/10.1038/cr.2011.145; PMID: 21876558
- Chang EC, Barr M, Wang Y, Jung V, Xu HP, Wigler MH. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 1994; 79:131 - 41; http://dx.doi.org/10.1016/0092-8674(94)90406-5; PMID: 7923372
- Onken B, Wiener H, Philips MR, Chang EC. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci U S A 2006; 103:9045 - 50; http://dx.doi.org/10.1073/pnas.0603318103; PMID: 16754851
- Chang EC, Philips MR. Spatial segregation of Ras signaling: new evidence from fission yeast. Cell Cycle 2006; 5:1936 - 9; http://dx.doi.org/10.4161/cc.5.17.3187; PMID: 16931912
- Cheng CM, Chang EC. Busy traveling Ras. Cell Cycle 2011; 10:1180 - 1; http://dx.doi.org/10.4161/cc.10.8.15259; PMID: 21436618
- Zheng ZY, Cheng CM, Fu XR, Chen LY, Xu L, Terrillon S, et al. CHMP6 and VPS4A mediate the recycling of Ras to the plasma membrane to promote growth factor signaling. Oncogene 2012; In press http://dx.doi.org/10.1038/onc.2011.607; PMID: 22231449
- Kerppola TK. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol 2008; 85:431 - 70; http://dx.doi.org/10.1016/S0091-679X(08)85019-4; PMID: 18155474
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009; 458:445 - 52; http://dx.doi.org/10.1038/nature07961; PMID: 19325624
- Baldys A, Raymond JR. Critical role of ESCRT machinery in EGFR recycling. Biochemistry 2009; 48:9321 - 3; http://dx.doi.org/10.1021/bi900865u; PMID: 19673488
- Lu A, Tebar F, Alvarez-Moya B, López-Alcalá C, Calvo M, Enrich C, et al. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J Cell Biol 2009; 184:863 - 79; http://dx.doi.org/10.1083/jcb.200807186; PMID: 19289794
- Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, et al. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal 2011; 4:ra13; http://dx.doi.org/10.1126/scisignal.2001518; PMID: 21386094
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011; 11:761 - 74; http://dx.doi.org/10.1038/nrc3106; PMID: 21993244