 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The early detection of invading viruses by the host depends on their identification by pathogen sensors. These include Toll-like receptors (TLRs) as well as cytoplasmic RNA helicases such as retinoic acid inducible protein I (RIG-I) and melanoma differentiation associated gene 5 (MDA-5). These pathogen sensors recognize specific molecular patterns found in viruses and trigger inflammatory and antiviral responses that result in the eradication of invading pathogens. In this study we investigated the specific recognition of Human rhinovirus 6 (HRV6) the common cold pathogen by the innate immune response in lung epithelial cells. Our experiments established that in the first stages on infection the TLRs play a crucial role in HRV recognition and that different constituents of HRV6 are recognized by different TLRs, while upon viral replication and generation of dsRNA the type I IFN inflammatory response is mediated by MDA-5. The HRV6 capsid is recognized via TLR2, whereas upon HRV6 ssRNA internalization the virus genome is recognized by TLR7 and TLR8. Upon generation of dsRNA the type I IFN response is mediated by MDA-5. The combined recognition by different TLRs and MDA5 and their upregulation concurs with the huge inflammatory response seen in the common cold caused by human rhinoviruses.
Introduction
Human rhinoviruses, the common cold pathogens, are small, non-enveloped single stranded RNA viruses, which affect a significant number of humans during the winter months. Human infection causes severe inflammation in the upper respiratory system, causing nasal secretions and cough. Although rhinovirus infections are not life-threatening, epidemiological studies have documented their involvement in acute respiratory illness in various human populations.Citation1,Citation2 Lower respiratory disease has also been reported among infected infants and children and rhinoviruses have been associated with bronchiolitis, chronic bronchitis and atypical pneumonia in children.Citation3,Citation4
Despite being one of the most frequent viral infections in humans, elucidation of the host defense mechanisms that are triggered by HRVs have not been fully resolved.
To date a number of viruses have been shown to trigger innate immune responses via Toll-like receptors. Toll-like receptors (TLRs) have a key role in the innate immune response. They are essential sensors of microbial pathogens. TLRs recognize specific molecular patterns found in microbial pathogens and trigger inflammatory and antiviral responses that result in the eradication of invading pathogens.Citation5,Citation6
Individual TLRs are differentially distributed within the cell and each TLR has the important role of recognizing specific microbial components derived from pathogens including bacteria, viruses, protozoa and fungi. Many TLRs have been shown to be involved in viral recognition; for example, TLR4 recognizes Syncytial virusCitation7 and Coxsackievirus B4.Citation8 TLR2 plays a key role in the pro-inflammatory response against various viruses including herpes simplex virus and cytomegalovirus.Citation9 While TLR3 has been shown to recognize viral components such as poly(I:C)Citation10 and dsRNA from Lang reovirus and West Nile virus.Citation11 TLR9 recognizes bacterial and viral CpG DNA motifs like CpG DNA motifs found in Herpes simplex virus (HSV).Citation12,Citation13 TLR7 and TLR8 mediate recognition of ssRNA and have been shown to recognize many ssRNA viruses such as Influenza virus,Citation14–Citation16 Coxsackievirus B3,Citation17 and Human Parechovirus 1.Citation18
A second group of pattern recognition receptors that “detect” invading viruses has been identified recently. These receptors are RIG-like receptors (RLRs) and include RIG-I (retinoic acid inducible protein I) and MDA5 (melanoma differentiation associated gene 5) and have been implicated in viral RNA recognition.Citation19–Citation21 RIG-I has been implicated in the innate recognition of Paramyxovirus together with TLR7 and TLR8.Citation22 While MDA5 triggers IFN responses to encephalomyocarditis virus.Citation23 Studies have revealed that RIG-I recognizes single-stranded RNA (ssRNA) containing a terminal 5-triphosphate (ppp),Citation24,Citation25 as well as linear dsRNA no longer than 23 nucleotides.Citation39,Citation42 MDA5 recognizes long strands of dsRNA but the mechanism by which this occurs is less clear.Citation39,Citation42
In this study we investigated the specific recognition of human rhinoviruses by the innate immune system. Our results showed that the massive inflammatory response triggered by these viruses is due to the specific recognition of different viral components by different pattern recognition receptors. Initially the innate immune antiviral response is mediated by different TLRs. We show that TLR2 can recognize HRV viral proteins, whereas once the virus is internalized the endosomal-located TLR7 and TLR8 are vital for the recognition of HRV ssRNA. Once the virus begins replication, dsRNA is generated and it is mediated by the cytosolic sensor MDA5 is responsible for its recognition.
Results
TLR expression in HRV infection.
Toll-like receptors recognize different pathogen-associated molecular patterns (PAMPs) leading to the activation of an innate immune response and the shaping of the subsequent adaptive immune response. In order to investigate the involvement of TLRs in human rhinovirus infection we examined the expression of different TLRs (TLR1, TLR2, TLR3, TLR4, TLR6, TLR7, TLR8) from bronchial epithelial cells before and after HRV6 infection. To establish if a particular constituent of the viral particle was responsible for the cellular response cells were incubated with live HRV6, UV-inactivated HRV6 (which is non-infectious) as well as HRV6 naked viral ssRNA for different time points up to a complete infectious cycle (0.5 h, 1 h, 2 h, 4 h, 6 h, 8 h).
Our data revealed that although TLR expression was minimal on these cells before infection there was a high increase in TLR2 expression with HRV6 and UV-inactivated virus ( and C). TLR7 and TLR8 showed increase with both the live virus and the naked ssRNA ( and B).
When we tested respiratory epithelial cells for RIG-I and MDA5 expression we found low expression. After the cells had been stimulated with live HRV6, UV-inactivated virus, and viral ssRNA for different time points there was a significant upregulation of MDA5 at 4 h in cells stimulated with live virus (). It is not surprising that the upregulation of MDA5 is observed at 4 h, since RNA synthesis for this virus begins at approximately 4–5 h post-infection.
Cytokine secretion is triggered by HRV6 infection.
In order to determine the immuno-stimulatory effect of the virus, cells were stimulated for different time points with live HRV6, UV-inactivated HRV6 (unable to infect the cells and replicate) as well as HRV6 ssRNA. Our experiments showed secretion of pro-inflammatory cytokines in airway epithelial cells under all stimuli. IL-6 and to a lesser degree IFNβ showed increased secretion when stimulated with live HRV6 and HRV6 ssRNA while UV-inactivated HRV6 stimulated cytokine secretion, but to a lesser degree. IL6 showed significant increase at 2 h and then began decreasing whereas IFNβ showed a marked increase at 4 h post infection which is consistent with the virus replication ().
To further determine the role of TLRs as well as RIG-I and MDA-5 in HRV6 recognition we used RNA interference to knock down expression of TLR2, TLR7, TLR8 or RIG-I and MDA-5 on respiratory epithelial cells. Once the knockdown was confirmed by western blotting, we stimulated the cells with HRV6, HRV6 ssRNA and UV-inactivated HRV6 and measured IL6 () and IFNβ secretion (). Our results showed that the TLRs contribute mainly to IL6 secretion, whereas MDA5 mainly contributes to IFNβ secretion. When HRV6 ssRNA was added to the knock down TLR7 and TLR8 cells a considerable decrease in cytokine secretion was observed. Subsequently, when TLR2 was silenced there was a decrease in cytokine secretion when cells were stimulated with HRV6 and UV-inactivated HRV6, thus demonstrating that TLR2 is the host sensor for the viral capsid. Knocking down RIG-I had no effect in cytokine secretion. Knocking down MDA5 had no effect in ssRNA or UV HRV6 recognition but when we stimulated with live HRV6 a considerable decrease in IFNβ secretion was observed, therefore confirming that MDA-5 activation requires HRV6 replication competence and production of dsRNA. Furthermore our results show that the IFNβ immune response to HRV6 in airway epithelial cells is mainly regulated by MDA5 and to a lesser extent by the TLRs.
NFκB activation.
TLRs act upstream of NFκB activation. TLR signalling pathways have been shown to ultimately result in the release of NFκB from its endogenous inhibitor and subsequent nuclear translocation that leads to the secretion of proinflammatory cytokines. In order to determine which receptors are responsible for the cytokine secretion that we observed when we used the different HRV6 we utilized human embryonic kidney (HEK) cells transfected with either TLR2, TLR3, TLR4, TLR6, TLR7, TLR8, MDA5, RIG-I and an NFκB luciferase reporter gene. Furthermore we used HEK cells transfected with TLRs, MDA5 or RIG-I and an IRF3 luciferase reporter gene.
All HEK/TLR cell lines were stimulated with HRV6 virions, HRV6 purified ssRNA, as well as UV-inactivated HRV6, in order to test whether cell activation was dependent on viral replication competence. In addition, all HEK transfected cells were stimulated with their appropriate ligands as a positive control, i.e., HEK/TLR2 cells were stimulated with LTA, while HEKTLR4/MD2/CD14 were stimulated with LPS. HEK/TLR3 cells stimulated with dsRNA from reovirus, HEK/TLR7 and HEK/TLR8 with GU rich RNA nucleotides, while HEK/MDA5 cells and HEK/RIG-I cells were stimulated with EMCV and with influenza virus respectivelyCitation33 ().
After 6 h of stimulation, the cells were lysed in passive lysis buffer (Promega). Luciferase activity was measured using a plate reader luminometer. All transfected cell lines responded adequately to their appropriate ligand thus verifying the successful recognition of respective known ligands.
Our experiments revealed that the viral genomic RNA is recognized through TLR7 and TLR8, since not only HRV6 virions but also HRV6 ssRNA activated NF.B expression on HEK/TLR7 and HEK/TLR8 cells. TLR7 and TLR8 together with TLR3 and TLR9 belong to a group of TLRs that recognize nucleic acids intracellularly. Single stranded RNA (ssRNA) from many different viruses has been shown to stimulate innate immune recognition via TLR7 and TLR8; thus it is not surprising that these two TLRs are responsive to HRV6 genomic RNA (). In addition, live HRV6 and UV-inactivated HRV6 produced cell activation in HEK/TLR2 cells. There was no activation of RIG-I but a significant activation of MDA5 with live HRV6 but no activation with HRV6 ssRNA or UV-inactivated virus ().
HRV6 molecular pattern confers recognition by TLR2.
Our results demonstrate that TLR2 recognizes HRV6 particles as well as UV-inactivated HRV6 particles but not HRV6 ssRNA, thus suggesting that the triggering of the inflammatory cytokines through TLR2 in response to HRV6 is independent of internalization and replication competence of the virus. Since UV-inactivated particles trigger inflammation via TLR2, the results indicate that the constituent responsible for the activation and upregulation of TLR2 must be the virus capsid.
In order to verify these findings HEK/TLR2 cells were preincubated with TLR2-specific mAb (TLR2.1) prior to incubation with HRV6 and UV-inactivated HRV6. The data showed that HRV6 and UV HRV6-induced cellular activation was inhibited by the TLR2 specific mAb (TLR2.1) (), thus suggesting the importance of TLR2 in HRV6-mediated activation of cells.
Single stranded HRV6 RNA recognition in endosomes.
It has been proposed that TLR3, TLR7, TLR8 and TLR9 are part of an isolated group within the TLR family that recognizes viral nucleic acids in the endosome or lysosome. Our results suggest that TLR7 and TLR8 are activated by HRV6 ssRNA. In order to determine whether HRV6 ssRNA engages TLR7 and TLR8 directly we utilized fluorescence resonance energy transfer (FRET).
FRET was measured in terms of dequenching of donor fluorescence after complete photo-bleaching of the acceptor fluorophore. Increased donor fluorescence after complete destruction of the acceptor indicated association between the two molecules of interest. Prior to our experiments, the energy transfer efficiency in our system was measured using a positive control (MHC-class I and β2-m) as well as a negative control for the energy transfer (see ). As expected maximum energy transfer efficiency (E) was 29 ± 1.2%.
In order to visualize the viral RNA, we labeled purified HRV6 ssRNA with Alexa 488 using the ULYSIS nucleic acid labeling kit. Although Picornavirus naked RNA has been shown in previous studies to be infectiousCitation27,Citation28 (at a much lower percentage than the complete virions) to cells, we also used cationic lipids to facilitate uptake of RNA, since it has been shown that RNA complexed with DOTAP can be easily uptaken by a cell.Citation29
We measured FRET on epithelial cells between TLR7, TLR8 or TLR3 (using Alexa 543-conjugated TLR-specific Fab mAb) and HRV6 ssRNA-Alexa 488. Our results demonstrated that TLR7 and TLR8 were associating with viral ssRNA, giving FRET values of 18 ± 1.6% and 24 ± 0.8% respectively (), whereas TLR3 was not associating with HRV6 ssRNA, giving FRET values of 5 ± 0.7%.
When polyIC-Alexa 488 was used as a control there was no association of TLR8 or TLR7 and poly I:C-Alexa 488 while we found increased association between TLR3 and polyIC-Alexa 488 (E = 27 ± 2.0%) (), thus verifying the specificity of our results.
The functional significance of endosomal recognition was also investigated by disrupting the endosomal pH in order to see whether we could affect the HRV6 ssRNA recognition. We used NH4Cl, which is a weak lysosomotropic base that diffuses into acidic endosomes, where it becomes protonated. Once protonated it is unable to diffuse out, thus increasing the pH. We found that cytokine secretion was inhibited when epithelial cells were treated with NH4Cl, prior to stimulation with HRV6 ssRNA, thus suggesting that endosomal integrity is crucial for viral RNA-induced recognition. The cells were treated with 25 nM or 50 nM NH4Cl, and FRET was used to determine associations. Our results demonstrate that there was a significant decrease in TLR8 and TLR7 associations with HRV6 ssRNA as well as a significant decrease in TLR3 and poly I:C interactions (). Furthermore, when IL-6 secretion was measured in response to ssRNA, it was also found to be significantly reduced in cells treated with NH4Cl ().
In order to rule out the possibility that the FRET observed was due to random distribution, we varied the ratio of donors and acceptors used to label the proteins of interest. E was found to be independent on acceptor density, to be sensitive to donor:acceptor ratio, and not to go to zero at low surface density, thus suggesting that the FRET values observed were due to clustered molecules and not random associations.
Discussion
Viruses over the years have developed a fascinating array of mechanisms to infect the host cells. Although these stealth mechanisms are essential for the preservation of some viruses, it is nearly impossible for a virus to enter a cell and set up housekeeping without setting off the alarm system that triggers the innate immune response.
The innate immune system is highly conserved and is the first line of defense for protecting the host from invading microbial pathogens. In the case of viruses, two families of pattern recognition receptors are involved in the innate immune sensing, the Toll-like receptors (TLRs) and the RIG-like receptors (RLRs). They are essential sensors of microbial pathogens.Citation5 TLRs recognize specific molecular patterns found in microbial pathogens and trigger inflammatory and antiviral responses that result in the eradication of invading pathogens.Citation34 RLRs recognize viral RNA independently of TLRs and unlike TLRs, which are found either on the cell surface or endosomes RLRs are found in the cytoplasm where cellular RNA is also present.Citation35 It has been shown that RIG-I and MDA5 recognize different viruses and different viral RNAsCitation36,Citation37 (). RIG-I recognizes single stranded RNA (ssRNA) containing a terminal 5-triphosphate (ppp),Citation38 as well as linear dsRNA no longer than 23 nucleotides.Citation39 MDA5 recognizes long strands of dsRNA but the mechanism by which this occurs is less clear.Citation39
In this study we investigated whether TLRs are involved in the inflammatory response that is induced by HRVs in human respiratory epithelial cells. Rhinoviruses are non-enveloped ssRNA viruses that can infect the upper and lower human respiratory system and cause the common cold but they are also associated with respiratory disease as well as bronchitis and atypical pneumonia.Citation4,Citation40
Our experiments established that TLRs play a crucial role in HRV recognition. Furthermore we showed that different constituents of HRV6 were recognized by different TLRs and that the HRV-induced inflammatory response by the host was due to a synergic effect mediated via TLR2, 7 and 8.
Our results suggest that TLR2 contributes to the inflammatory response by both live and UV-inactivated virus particles. TLR2 is expressed on the cell surface of human respiratory cells and does not require replication competence of HRV6. Therefore it seems that TLR2 recognizes and responds to a particular motif or molecular pattern on the virus capsid.
TLR7 and TLR8 have been shown by previous studies to be the “key sensors” of viral ssRNA.Citation14,Citation18 They reside in the endosomes and can recognize ssRNA from different viruses; thus it is not surprising that they sense HRV6 genomic ssRNA and mount an innate immune response. Our FRET studies confirmed the association of ssRNA HRV6 with TLR7 and TLR8.
The involvement of TLR3 in HRV6 recognition was also investigated. A study by Hewson et al.Citation41 has shown that TLR3 mediates rhinovirus activation, but in our study we did not see any TLR3-induced HRV6 recognition. One of the reasons could be that we are using different cell types as well as different rhinovirus serotypes and they could differ at the activation of innate immune recognition.
The involvement of RLRs was also examined. Our data are in agreement with Kato et al.Citation33 that MDA5 and RIG-I have differential roles in the recognition of RNA viruses. We have seen that MDA5, but not RIG-I, is implicated in the recognition of rhinoviruses and the subsequent production of type I interferons. MDA5 is activated later than the TLRs, probably recognizing the dsRNA generated upon viral replication.
Overall our data suggests that HRV6 like many other viruses, such as HSV and CMV is sensed by more than one PRR. TLR2, TLR7 and TLR8 are heavily involved in HRV6 recognition. In addition MDA5 seems to be involved in HRV6 recognition subsequent to TLRs once viral replication begins generating dsRNA. The combined recognition of all these TLRs and MDA5 and their upregulation concurs with the huge inflammatory response seen in the common cold. It seems that the particular TLR transmitting the “invasion signal” might be dependent on the means of virus entry into the cell and the properties of the infected cell. We have seen that in the case of HRV6, viral capsid proteins trigger a TLR2-based “cell surface sensing mechanism” that begins the pro-inflammatory antiviral response. As the virus enters the cell the virus genome is detected by the localized endosomal TLR7 and TLR8, which are ideally situated for detecting viral nucleic acids. The use of inhibitors of endosomal acidification such as NH4Cl prevents recognition of the virus genome verifying the importance of these TLRs in innate responses. Once the virus enters the host and begins replication MDA5 senses the dsRNA produced and continues the pro-inflammatory antiviral response.
In conclusion we believe that it is the immune system's synergistic innate recognition of rhinoviruses by several TLRs and MDA5, which results in an overzealous pro-inflammatory antiviral response, and the exaggerated symptoms of the common cold.
Materials and Methods
Cell lines.
Human embryonic kidney 293 (HEK-239) cells transfected with TLR2, TLR3, were maintained in DMEM containing 4.6 g/L glucose with 10% FCS, 10 µg/ml ciprofloxacin and 0.5 mg/ml G418-sulfate. HEK-TLR4/MD2/CD14 (Visintin, 2003 15/id) was kindly provided by Professor Douglas Golenbock (University of Massachusetts). Transfections of HEK/TLR7 and HEK/TLR8 were maintained in DMEM containing 4.6 g/L glucose with 10% FCS and 10 µg/ml blasticidin. Transfections of HEK/MDA5 and HEK/RIG-I cells with puno-hMDA-5, or puno-RIG-I (Invivogen) were performed using Lipofectamine 2000 according to the manufacturer's recommendations.
Primary human bronchial epithelial cells were obtained from an adult male (TCS cell works) and maintained in the epithelial cell medium provided from TCS. The cells were grown in 25 cm2 easyflasks (nunc) and maintained at 37°C and 5% CO2. Primary cells were used between five and ten passages.
Virus.
Human rhinovirus 6 was obtained from the ATCC and propagated in human bronchial epithelial cells. The virus was purified in sucrose gradients as described in previous studies.Citation26 The infectivity of purified virus was determined by plaque titration.
UV-inactivated HRV6 was obtained by exposure to UV light at 1,200 mJ/cm2 for 30 min. To test viral inactivation, cells were exposed to UV HRV6 and the cytopathic effects were assessed.
Viral genomic RNA was isolated from purified virus using TRIzol Reagents (Invitrogen) according to the manufacturer's instructions. Viral purified RNA was used to stimulate cells at 25 µg/ml. Cationic lipids were used to facilitate uptake of RNA, since it has been shown that RNA complexed with DOTAP can be easily up-taken by a cell.Citation29 Fugene 6 (Roche, UK) to facilitate uptake of RNA was also used for RIG and MDA5 experiments, since it forms micelles that are taken up by cells and released in the cytoplasm.
Materials.
All fine chemicals were obtained from Sigma (UK). TLR2, TLR4 specific mAbs, were obtained from Hycult (Denmark). TLR3, TLR9, TLR7 and TLR8 specific antibodies were purchased from Santa Cruz Biotechnology (USA). MDA5 and RIG-I specific antibodies were purchased from Abcam (UK). Alexa 488-Ulysis reagent was obtained from Molecular Probes Inc. (UK). Single stranded RNA was labeled with Alexa-488-Ulysis reagent according to the manufacturer's instructions.
Luciferase reporter assays.
Cells (2 × 105) were transfected by using Lipofectamine-2000 according to the manufacturer's instructions with 200 ng of pNFκB-MetLuc2 (for NFκB activation) or 200 ng of 561 Luc (for IRF3 activation). Luciferase was determined by using the luciferase assay system reagents from Promega.
Statistical analysis.
Statistical analyses were performed using SPSS software version 15.0 (SPSS Inc., Chicago, USA). Comparison between two sets of patients or data was performed by the t-test or the Mann-Whitney U test.
RNA interference.
Transfections of siRNas were carried out using Oligofectamine. The target sequence of siRNA used were: for RIG-I, GGA AGA GGT GCA GTA TAT T, for MDA-5, GGT GAA GGA GCA GAT TCA G, for TLR2 GTC AAT TCA GAA CGT AAG TCA, for TLR7 GGG TAT CAG CGT CTA ATA TCA, for TLR8 GAC CAA CTT CGA TAC CTA AA.
Transfections with the specific siRNAs resulted in an approximately 60% decrease in receptor expression as determined by western blotting whereas transfection of cells with the scrambled siRNA did not show any decrease in TLR or RIG-I and MDA-5 expression.
Flow cytometric determination of TLR, RIG-I and MDA-5 expression.
In order to investigate TLR expression before and after HRV6 infection, human airway epithelial cells were either infected with HRV6 for 2 h or not, prior to fixation with 4% paraformaldehyde. The cells were subsequently washed and permeabilised using PBS/0.02% BSA/0.02% Saponin. After permeabilisation, the cells were incubated with antibodies against different TLRs and the appropriate secondaries conjugated to FITC. The cells were washed twice in PBS/0.02% BSA/0.02% Saponin and re-suspended in 500 µl of PBS. Fluorescence was detected using a FACSCalibur counting 10,000 cells not gated.
Fluorescence recovery after photobleaching (FRET).
FRET is a non-invasive imaging technique used to determine molecular proximity. FRET can occur over 1–10 nm distances, and effectively increases the resolution of light microscopy to the molecular level. It involves non-radiative transfer of energy from the excited state of a donor molecule to an appropriate acceptor. The rate of energy transfer is inversely proportional to the sixth power of the distance, between donor and acceptor. The efficiency of energy transfer (E) is defined with respect to r and R0, the characteristic Forster distance by:
In the present study, FRET was measured using a method as previously described.Citation30–Citation32 Briefly, samples were labeled with donor and acceptor conjugated antibodies and energy transfer was detected as an increase in donor fluorescence (dequenching) after complete photo-bleaching of the acceptor molecule.
Cytokine assays.
Cells used were un-stimulated or stimulated with HRV6 virions (1 × 103 PFU/ml) or ssRNA (25 µg/ml). The cultures were incubated for the designated times. The supernatants were collected and frozen until the cytokine assays were performed. The BectonDickinson cytometric bead array (CBA) systems were used in order to determine the level of multiple cytokines at the same time. The CBA system employs a series of beads with discrete fluorescence intensities that enable us to simultaneously detect multiple soluble cytokines. Each bead provides a capture surface for a cytokine and is analogous to a coated well in ELISA. In this study Human cytokine CBA Kits were used. We measured quantitatively Interleukin-2 (IL-2), Interleukin-4 (IL-4), Interleukin-6 (IL-6), Interleukin-10 (IL-10), tumor necrosis factor-α (TNFα) and Interferon-β (IFNβ) levels from each single sample.
Figures and Tables
Figure 1 TLR/helicase expression in human airway epithelial cells. Primary human bronchial cells were either not stimulated (0 h) or stimulated at different time points with HRV6 (1 × 103 PFU/ml) (A) HRV6 ssRNA (B) or UV HRV6 (C). The cells were fixed and permeabilized, followed by antibody staining against the particular receptor molecule and incubation with the appropriate secondary antibody conjugated to FITC. Fluorescence was detected using a FACSC alibur (BectonDickinson). The data presented are the mean of three independent experiments.
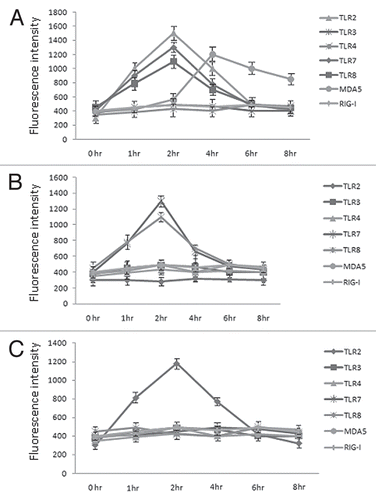
Figure 2 HRV6 activation of human airway epithelial cells. Human bronchial cells were either not stimulated (grey bar charts) or stimulated with HRV6 virions (1 × 103 PFU/ml) (black bar charts), UV HRV6 (white bar charts) or HRV6 ssRNA (stripped bar charts) for different hours. The supernatants were harvested and assayed for cytokine secretion using the Cytometric Bead Array (CBA) system (Becton Dickinson). Fluorescence was detected using a FACSC alibur (BectonDickinson). IL6 secretion is depicted in graph (A) while IFNβ in graph (B). The data presented is the mean of three independent experiments. Asterisks denote statistical significance (p < 0.001).
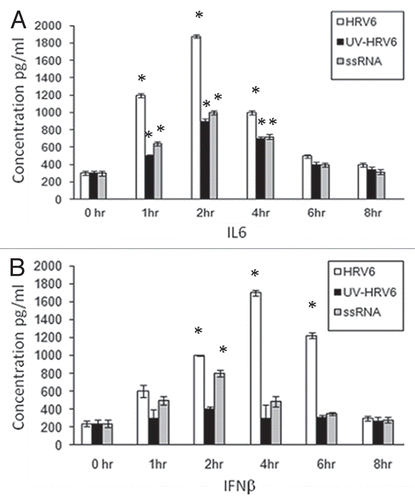
Figure 3 Inhibition of HRV6 activation of human airway cells by silencing TLRs MDA-5 and RIG-I. Following RNA interference (receptor expression levels were reduced by 80% by RNA interference A) human airway cells were either not stimulated (grey bar charts), incubated with HRV6 (black bar charts), incubated with UV-inactivated HRV6 (white bar charts) and HRV6 ssRNA (stripped bar charts). The supernatants were harvested and assayed for IL6 cytokine secretion in 2 h (B) and IFNβ secretion in 4 h (C) using the Cytometric Bead Array (CBA) system (Becton Dickinson). Fluorescence was detected using a FACSC alibur (BectonDickinson). The data represents the mean of three independent experiments. Asterisks denote statistical significance (p < 0.001).
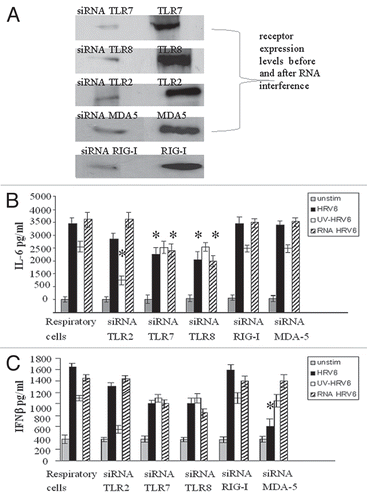
Figure 4 Receptor dependent activation in response to HRV6. HEK293 cells transfected with either TLR2, TLR3, TLR4, TLR7, TLR8, MDA-5 and RIG-I and a luciferase reporter gene were incubated with either no stimulus (grey bar charts), HRV6 virions (1 × 103 PFU/ml) (black bar charts), UV-inactivated HRV6 (white bar charts), ssRNA HRV6 (stripped black charts), specific receptor activating ligands (black dot charts). After stimulation, the cells were lysed and analyzed for luciferase activity. The data shown represent a mean of three independent experiments. Asterisks denote statistical significance (p < 0.001).
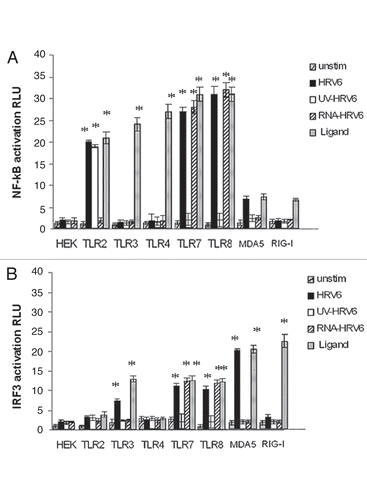
Figure 5 HRV6 recognition by TLR2. HEK293 cells transfected with TLR2 and a luciferase reporter gene were pre-incubated with TLR2-specific mAb (TLR2.1) prior to incubation with HRV6 (white bar charts) and UV HRV6 (black bar charts) (A). The cells were also incubated with an isotype control IgG2a from BD Biosciences (B). To determine NFκB activation the cells were lysed and analyzed for luciferase activity. The data shown represent a mean of three independent experiments. Asterisks denote statistical significance (p < 0.001).
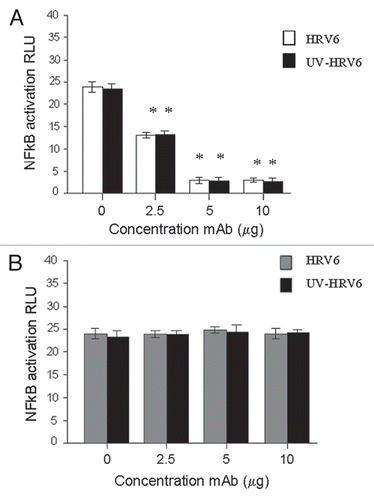
Figure 6 HRV6 ssRNA induced IL-6 secretion. HEK293 cells transfected with TLR7, TLR8 were either stimulated with HRV6 ssRNA (25 µg/ml) for 1 hour (stripped bar charts) or treated with 50 nM NH4Cl and then stimulated with HRV6 ssRNA (black bar charts). The supernatants were harvested and assayed for cytokine secretion using the Cytometric Bead Array (CBA) system (Becton Dickinson). Fluorescence was detected using a FACSCalibur (BectonDickinson). The data presented is the mean of three independent experiments. Asterisks denote statistical significance (p < 0.001).
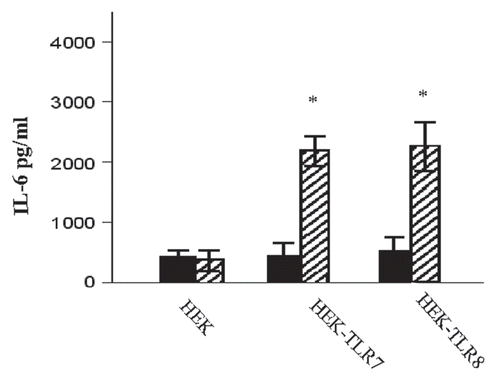
Table 1 Energy transfer efficiency values between donor-acceptor pairs
References
- Seemungal T, Harper-Owen R, Bhowmik A, Moric I, Sanderson G, Message S, et al. Respiratory viruses, symptoms and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001; 164:1618 - 1623
- Micillo E, D'Auria D, Mazzarella G, Abbate GF. Respiratory infections and asthma. Allergy 2000; 55:42 - 45
- Tsolia MN, Psarras S, Bossios A, Audi H, Paldanius M, Gourgiotis D, et al. Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis 2004; 39:681 - 686
- Hayden FG. Rhinovirus and the lower respiratory tract. Rev Med Virol 2004; 14:17 - 31
- Akira S. Toll-like receptors and innate immunity. Adv Immunol 2001; 78:1 - 56
- Akira S. Hemmi H. recognition of pathogen associated molecular patterns by TLR family. Immunol Lett 2003; 85:85 - 95
- Kurt-Jones EA, Popova L, Kwinn L, Haynes M, Jones LP, Tripp RA, et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytia virus. Nat Immunol 2000; 1:398 - 401
- Triantafilou K, Triantafilou M. Coxsackievirus B4-induced cytokine production in pancreatic cells is mediated through TLR4. J Virol 2004; 78:11313 - 11320
- Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, Golenbock DT, Finberg RW. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like Receptor 2. J Virol 2003; 77:4588 - 4596
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NFkappaB by Toll-like receptor 3. Nature 2001; 413:732 - 738
- Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med 2004; 10:1366 - 1373
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. TLR9 mediated recognition of Herpes Simplex 2 by plasmacytoid dendritic cells. J Exp Med 2003; 198:513 - 520
- Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, et al. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci USA 2001; 98:9237 - 9242
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of single stranded RNA viruses by Toll like receptor 7. Proc Natl Acad Sc USA 2004; 101:5598 - 5603
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004; 303:1529 - 1531
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species specific recognition of single stranded RNA via Toll like receptor 7 and 8. Science 2004; 303:1526 - 1529
- Triantafilou K, Orthopoulos G, Vakakis E, Ahmed MAE, Golenbock DT, Lepper PM, et al. Human cardiac inflammatory responses triggered by Coxsackie B Viruses are mainly Toll-like receptor (TLR) 8-dependent. Cellular Microbiol 2005; 7:1117 - 1126
- Triantafilou K, Vakakis E, Orthopoulos G, Schumann E, Lepper PM, Triantafilou M. TLR8 and TLR7 are involved in the host's immune response to Human Parechovirus 1. Eur J Immunol 2005; 35:2416 - 2423
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005; 23:19 - 28
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5 and LGP2 in antiviral innate immunity. J Immunol 2005; 175:2851 - 2858
- Johnson CL, Gale M Jr.. CARD games between virus and host get a new player. Trends in Immunol 2006; 27:1 - 4
- Melchjorsen J, Jensen SB, Malmgaard L, Rasmussenm SB, Weber F, Bowie AG, et al. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol 2005; 79:12944 - 12951
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA 2006; 103:8459 - 8464
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 2006; 314:997
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science 2006; 314:994
- Abraham G, Colonno RJ. Many rhinovirus serotypes share the same cellular receptor. J Virol 1984; 51:340 - 345
- Alexander HE, Koch G, Mountain IM, Sprunt K, Van Damme O. Infectivity of ribonucleic acid of poliovirus on Hela cell monolayers. Virology 1958; 5:172 - 173
- Smull CE, Ludwig EH. Infectivity of Poliovirus and its nucleic acid for dehydrated Hela Cell monlayers. J Bacteriol 1965; 89:52 - 57
- Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic cells pulsed with RNA are potent antigen presenting cells in vitro and in vivo. J Exp Med 1996; 184:465 - 472
- Bastiaens PI, Jovin TM. Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta1. Proc Natl Acad Sci USA 1996; 93:8407 - 8412
- Kenworthy AK, Edidin M. Imaging fluorescence resonance energy transfer as probe of membrane organisation and molecular associations of GPI-anchored proteins. Methods Mol Biol 1999; 116:37 - 49
- Triantafilou K, Fradelizi D, Wilson KM, Triantafilou M. GRP78 a co-receptor for Coxsackievirus A9, interacts with MHC-class-I molecules which mediate virus internalisation. J Virol 2002; 76:633 - 643
- Kato H, Takeuchi O, Sato S, Yoneyama M, Matsui K, Uematsu S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006; 441:101 - 105
- Kaisho T, Akira S. Toll-like receptor function and signaling. J Allergy Clin Immunol 2006; 117:979 - 987
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev 2009; 227:75 - 86
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006; 441:101 - 105
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, et al. Distinct Rig-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 2008; 82:335 - 345
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 2007; 314:997 - 1001
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 2008; 205:1601 - 1610
- Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res 2002; 85:41 - 46
- Hewson CA, Jardine A, Edwards MR, Laza-Stanca V, Johnston SL. Toll-like receptor 3 is induced by and mediates antiviral activity against rhinovirus infection of human bronchial epithelial cells. J Virol 2005; 79:12273 - 12279
- Saito T, Gale M Jr.. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med 2008; 205:1523 - 1527